1. What information is avaiable in eRNAbase?
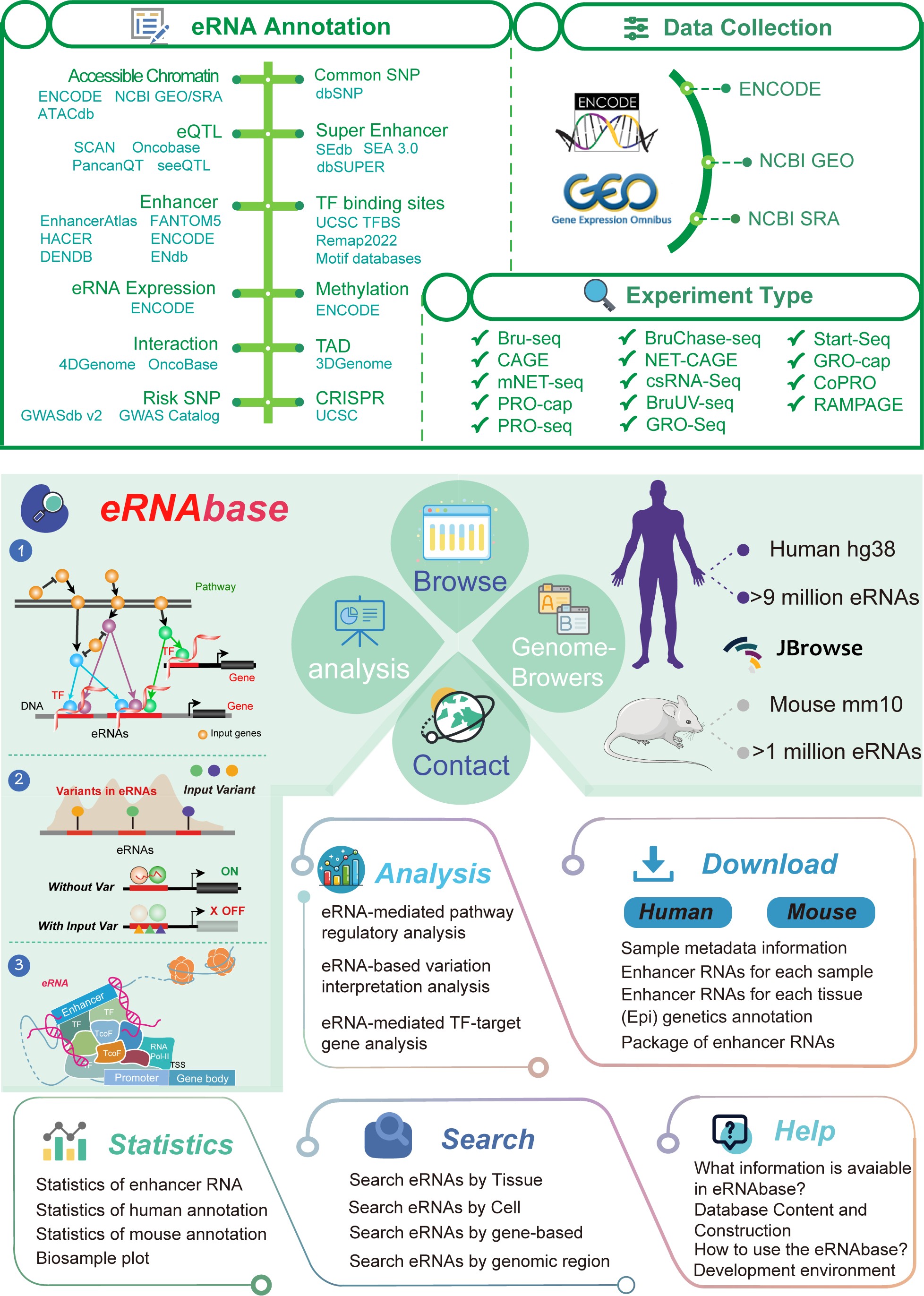
Enhancer RNA (eRNA) is a type of transcribed RNA from distal enhancers, which plays key roles in transcription regulatory circuitry to mediate the activation of target gene. Here, we developed the eRNA database (eRNAbase, https://bio.liclab.net/eRNAbase/index.php), which aims to document a large number of available resources of human and mouse eRNAs, provide comprehensive annotation and analyses for eRNAs.
2. What data are included in eRNAbase?
The current version of eRNAbase cataloged a total of 10, 421, 086 eRNAs from 1, 012 samples including 858 human samples and 154 mouse samples. These datasets were manually curated from numerous of epigenetic databases and covered 14 eRNA-related experiment types, including Bru-seq, CAGE, mNET-seq, PRO-cap, PRO-seq, BruChase-seq, NET-CAGE, csRNA-seq, BruUV-seq, GRO-seq, GRO-cap, Start-seq, CoPRO and RAMPAGE.
3. Database Content and Construction
eRNAbase provides the detailed and abundant (epi) genetic annotations in ChIP-seq based eRNA regions, such as super enhancer, enhancer, common SNPs, motif changes, expression quantitative trait locus (eQTL), risk SNPs, transcription factor binding sites (TFBSs), CRISPR/Cas9 target sites, DNase I hypersensitivity sites (DHSs), chromatin accessibility regions, methylation sites, chromatin interactions regions, TADs, and RNA spatial interactions. eRNAbase also provides eRNA-associated downstream target genes by mapping eRNA regions into genomes via five methods. Furthermore, eRNAbase provides three types of eRNA regulatory analyses for users, including eRNA-mediated pathway regulatory analysis, eRNA-based variation interpretation analysis and eRNA-mediated TF-target gene analysis. eRNAbase is a user-friendly database to query, browse and visualize information associated with eRNAs. We believe that eRNAbase could become a useful and effective platform for exploring potential functions and regulation of eRNAs in diseases and biological processes.
4. How to use the eRNAbase?
URL:https://bio.liclab.net/eRNAbase/browse.php
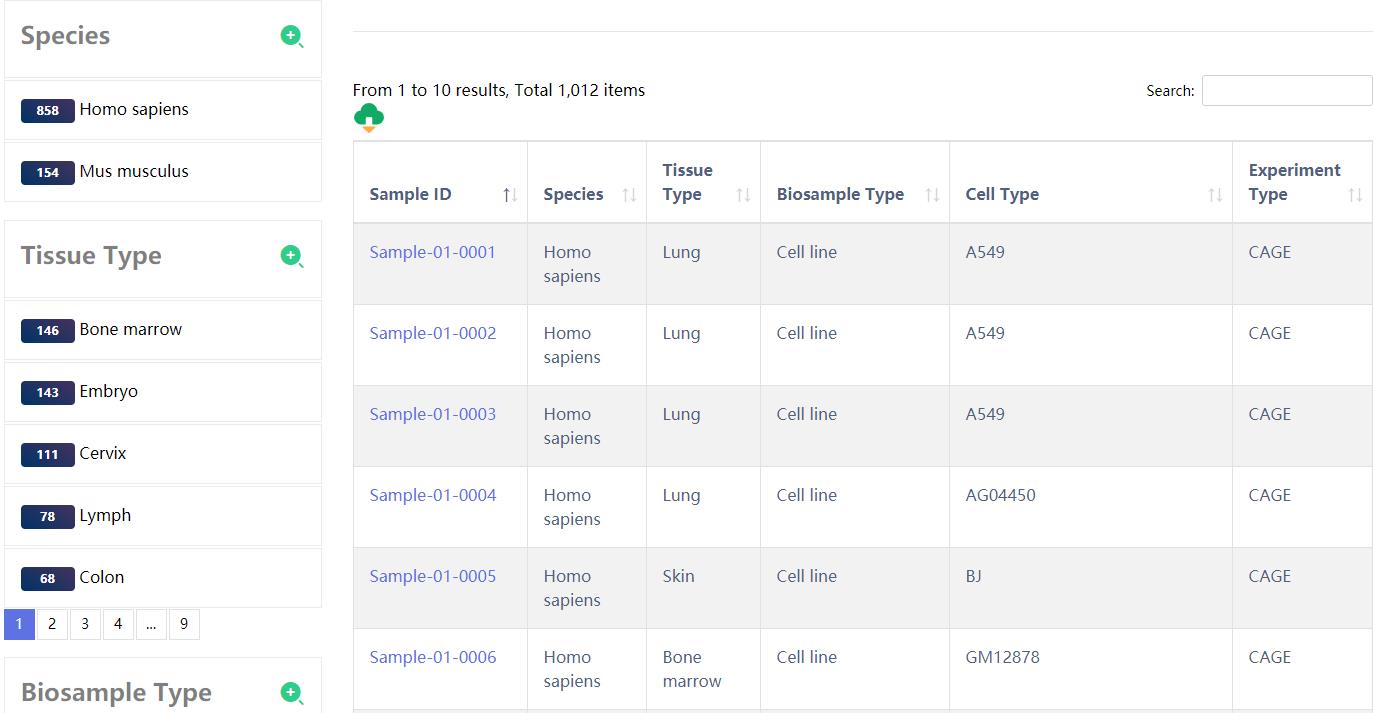
4.1 Browse
The 'Data-Browse' page is organized as an interactive and alphanumerically sortable table that allows users to quickly browse samples and customize filters through 'Species', 'Tissue type', 'Biosample Type', 'Cell Type' and 'Experiment type'. Users may further click on the 'Sample ID' to view eRNAs for a given sample.
URL:https://bio.liclab.net/eRNAbase/search.php
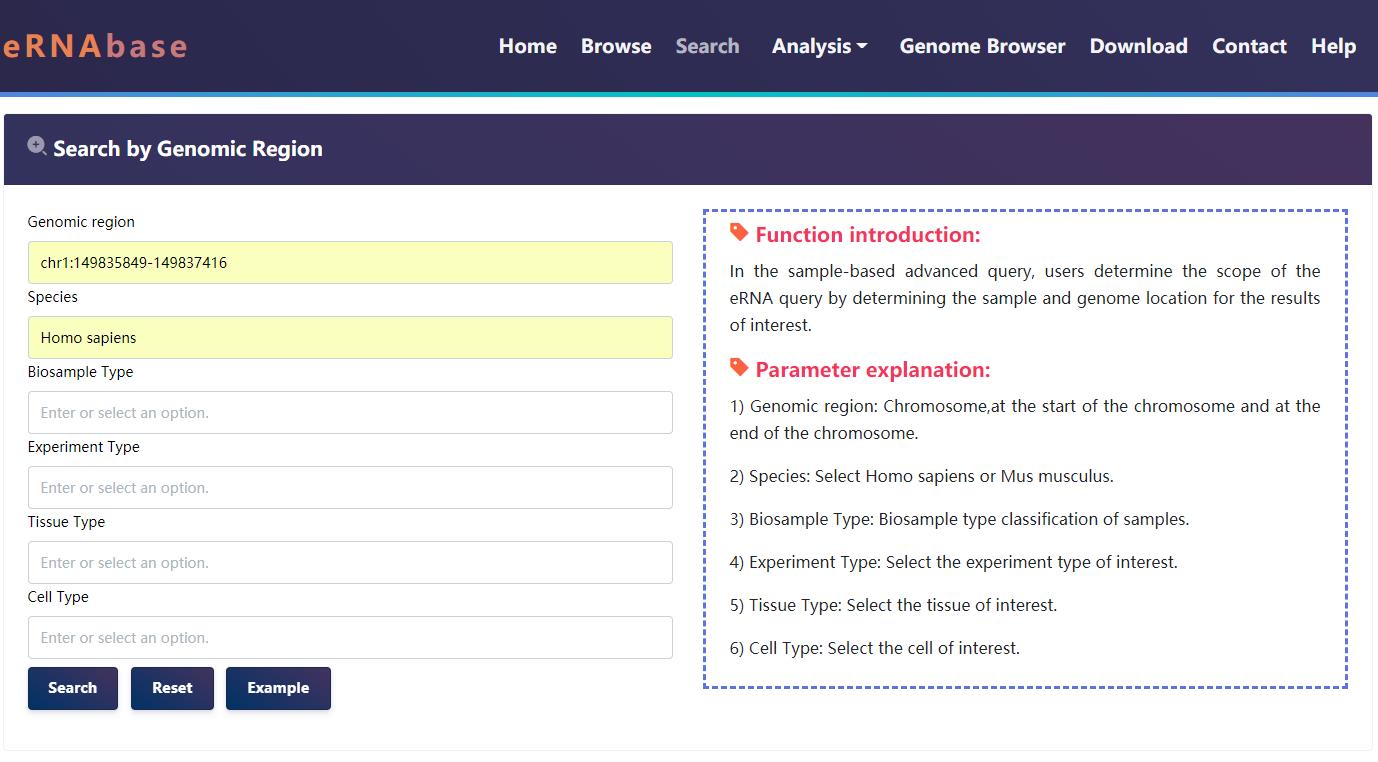
4.2 Search
eRNAbase provides four query search methods for users to obtain the eRNA data, including 'Search by genomic region' (input genomic position and select species, tissue/cell type and experiment type), 'Search by Target Genes' (select species, methods and input gene symbols of interest), 'Search by Tissue Type' (select species and input tissue of interest), and 'Search by Cell Type' (select species and input cell type of interest).
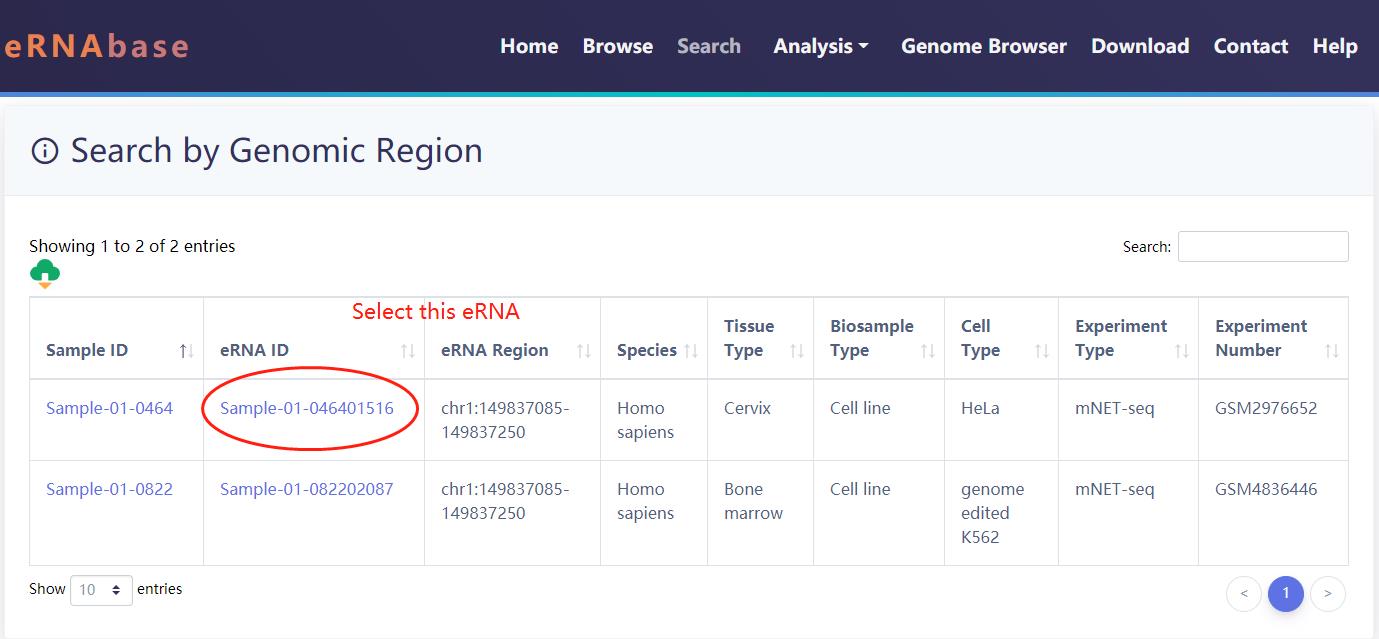
Example of search results for the human genomic region 'chr1:149835849-149837416'.
Users can select the eRNAs located in the genomic regions of interest.
eRNA overview information
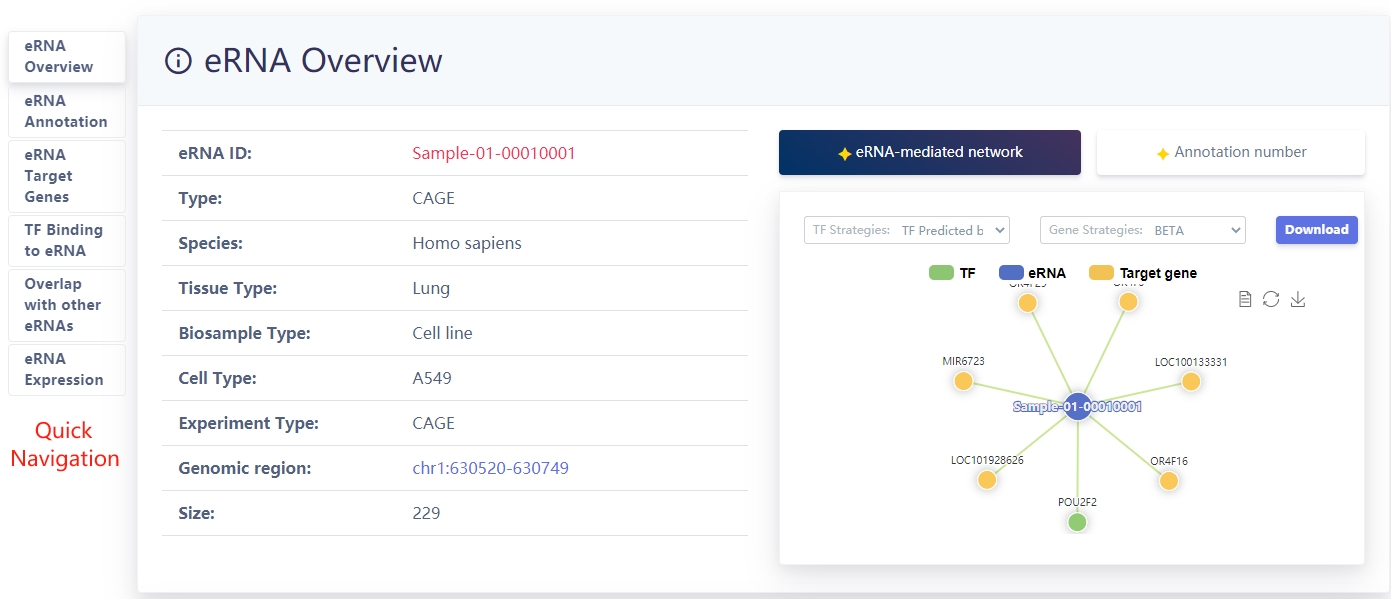
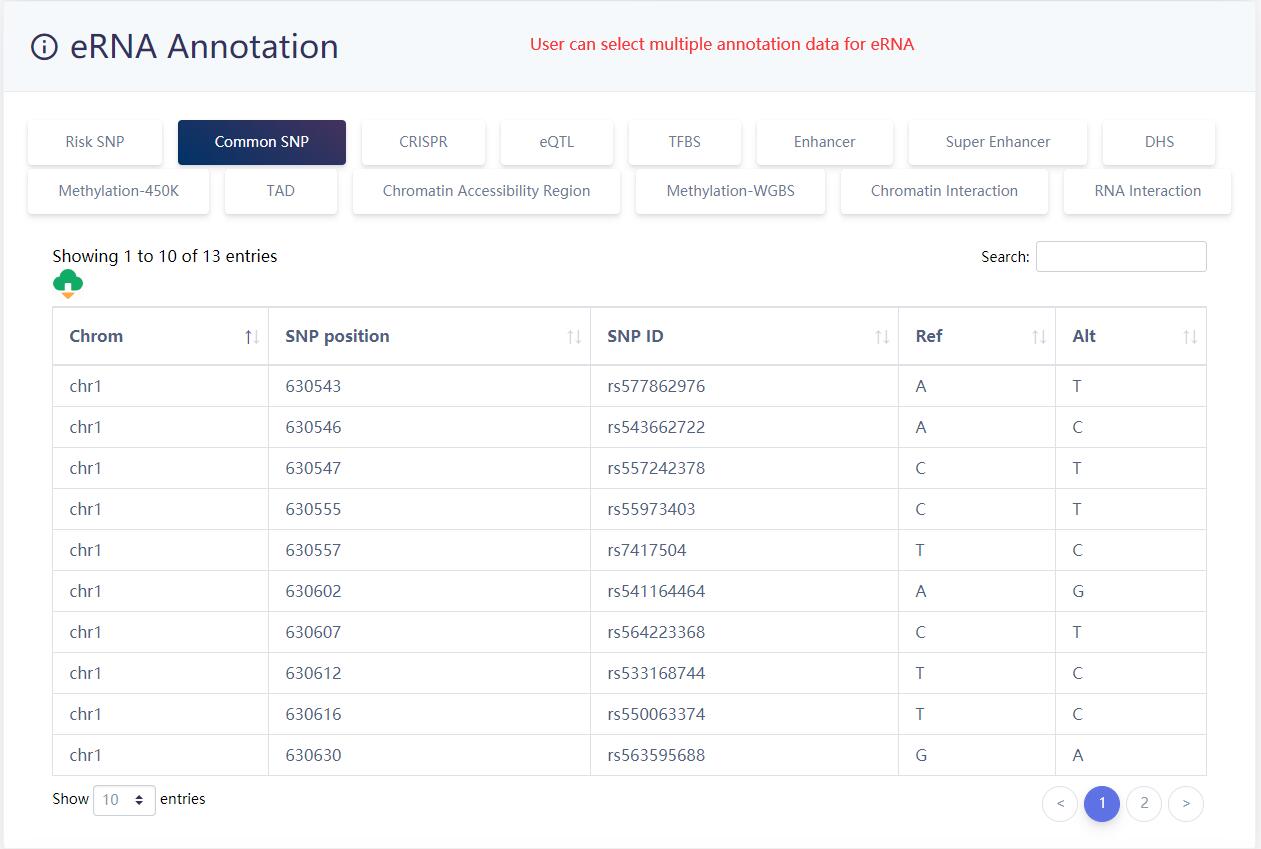
Users can select muitiple (epi)genetics annotation for eRNA.
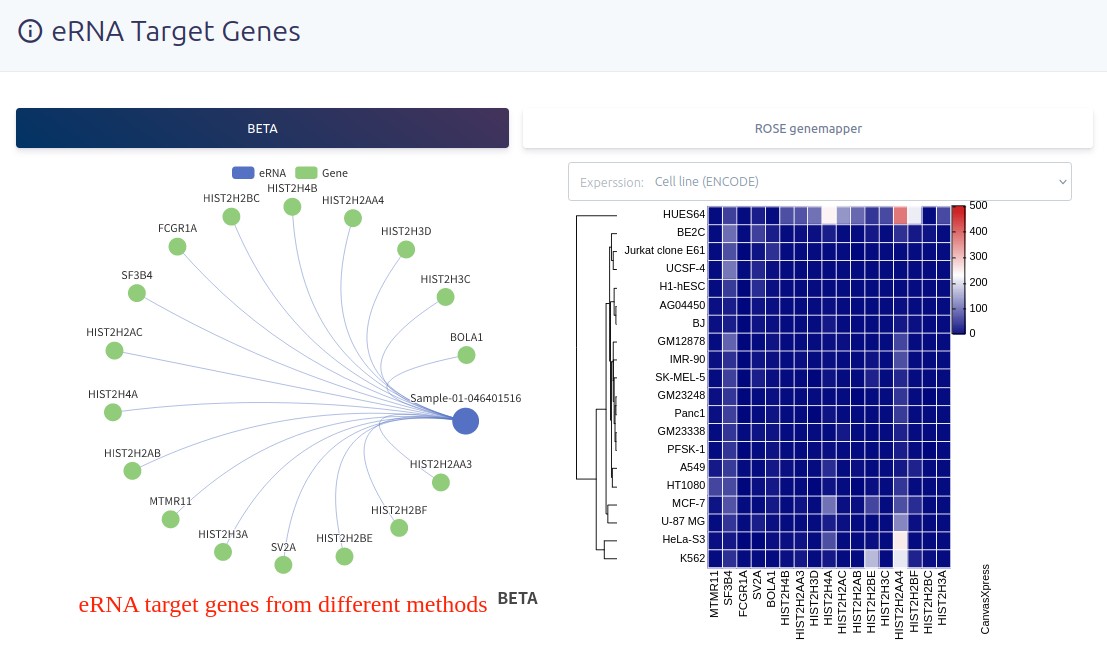
Users can choose the different methods to display the eRNA target genes.
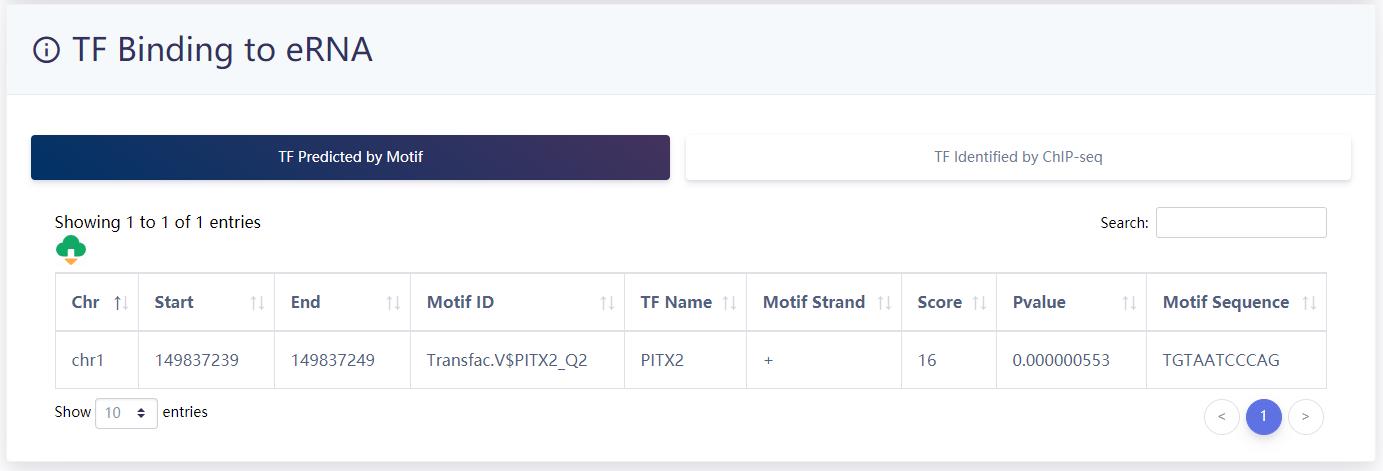
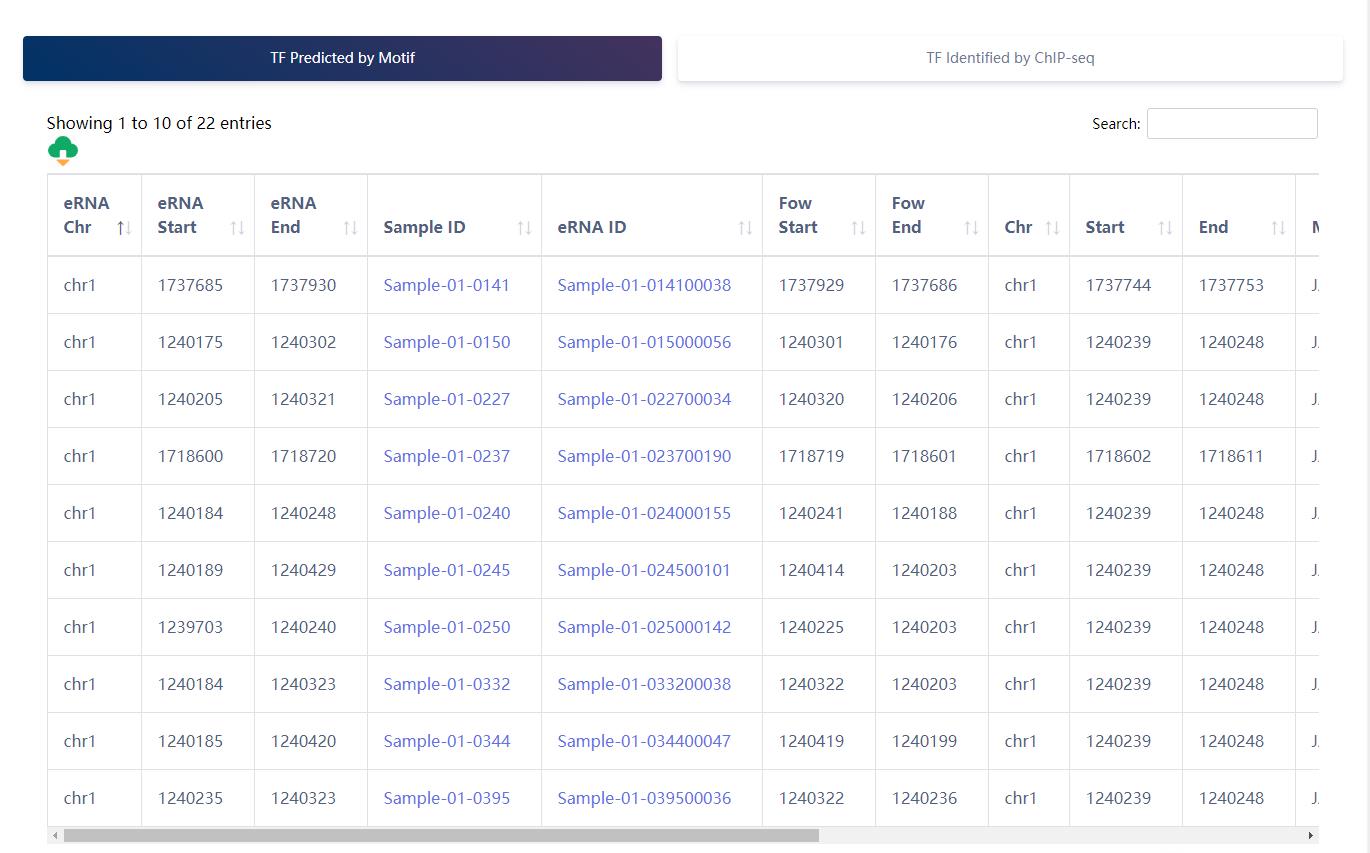
TF binding information of eRNA.
4.3 Analysis
URL:https://bio.liclab.net/eRNAbase/analysis/Pathway_downstream_analysis.php
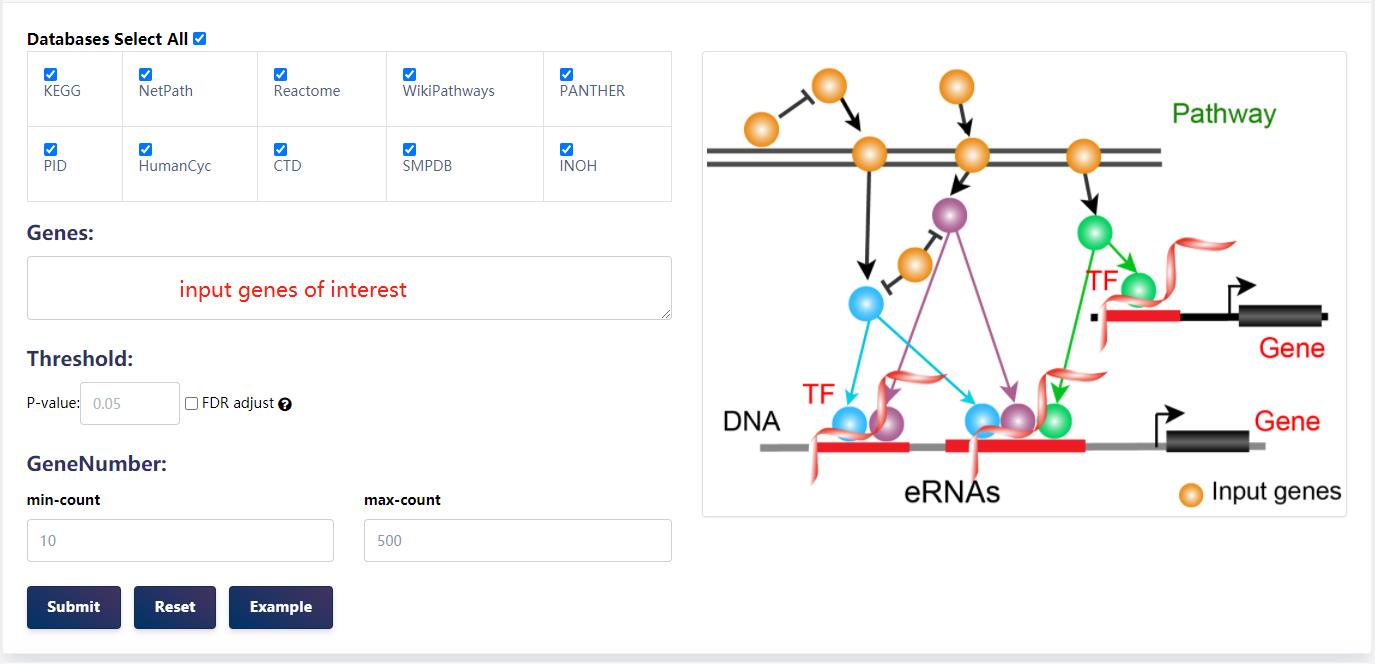
4.3.1 eRNA-mediated pathway regulatory analysis
Users can submit a gene list, choose the species (human or mouse), select at least one pathway database, set p-value/FDR and set gene number thresholds. eRNAbase will identify enriched pathways in up to the select pathway databases. In each pathway, eRNAbase will locate the terminal TFs and extract the terminal TF bound downstream eRNAs. Therefore, users can find the pathway/TFs/eRNAs/target genes regulatory axes.
The results of the analysis of the example data are as follows.
Users can select the enriched pathways of interest.
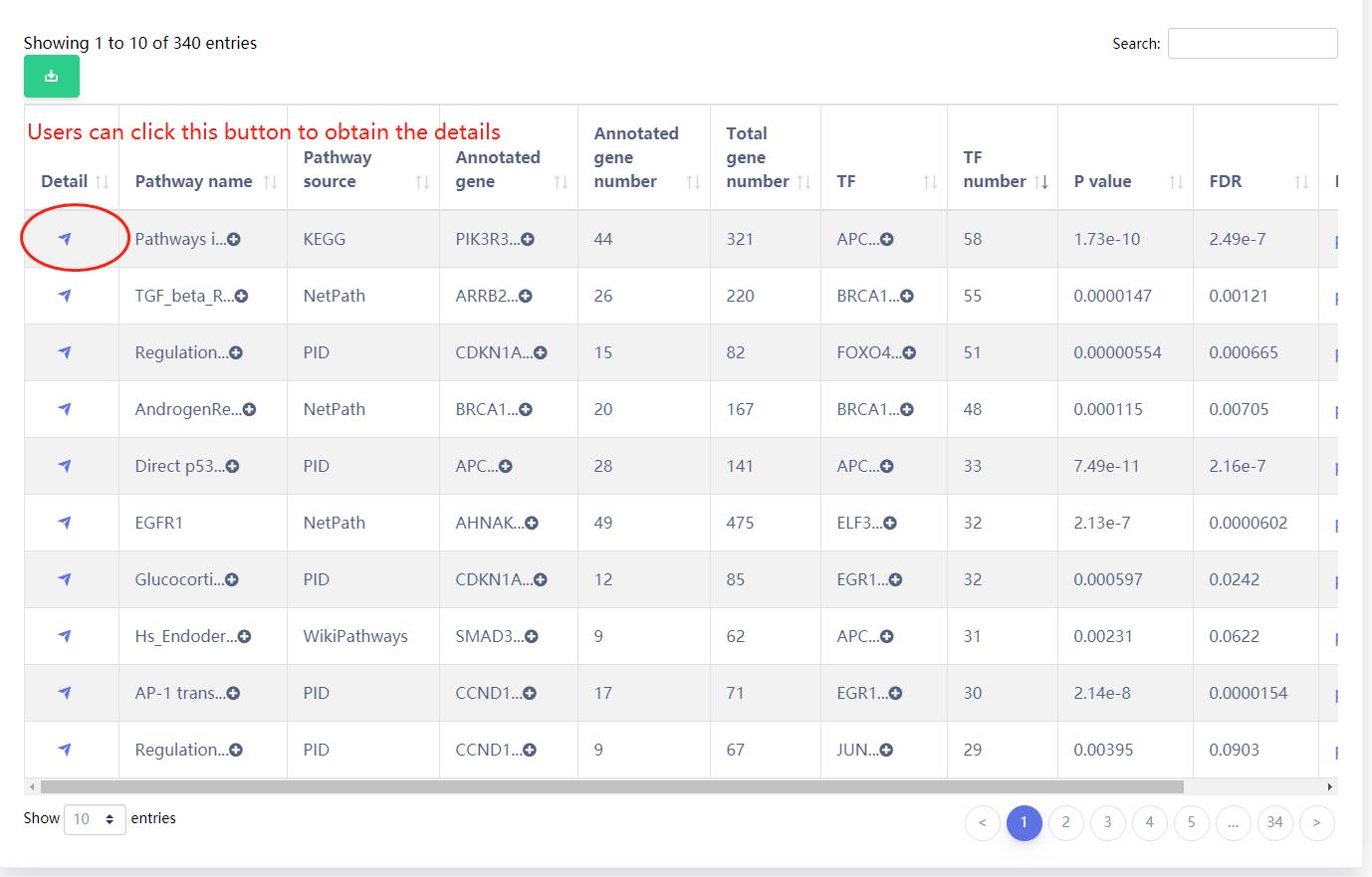
Pathway overview.

Pathway terminal TFs related eRNAs
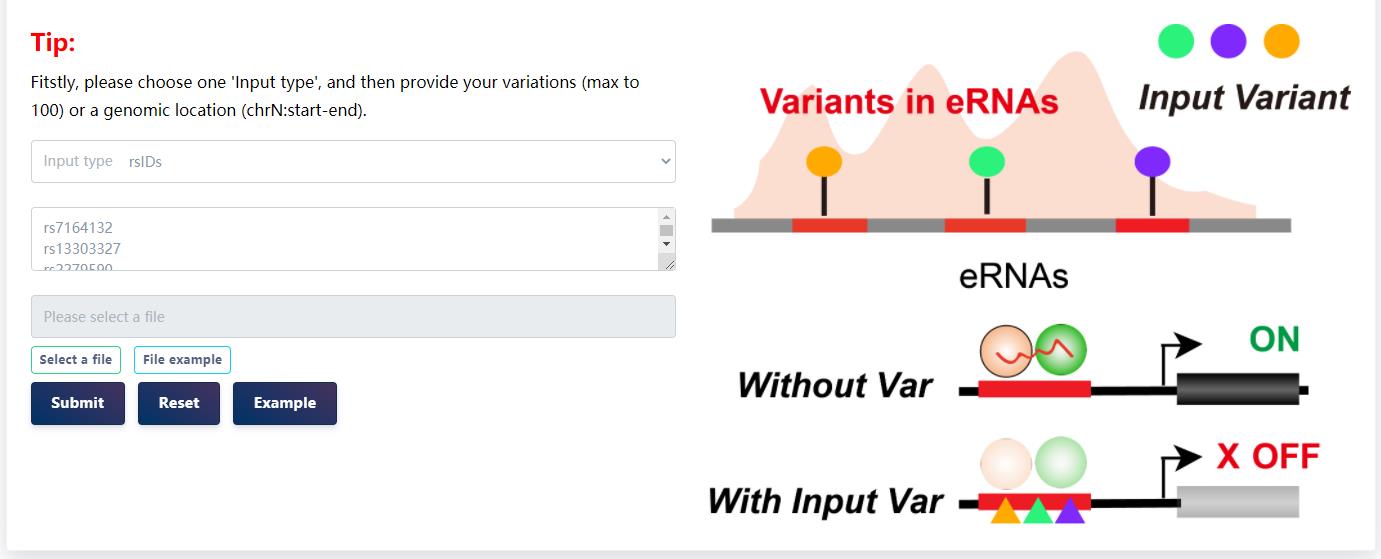
URL:https://bio.liclab.net/eRNAbase/analysis/eRNA_SNP.php
4.3.2 eRNA-based variation interpretation analysis
Users can submit a variant name list (rsIDs or genomic region) to eRNAs to extract the eRNAs that locate the variant.
The results of the analysis of the example data are as follows.
Variation overview.
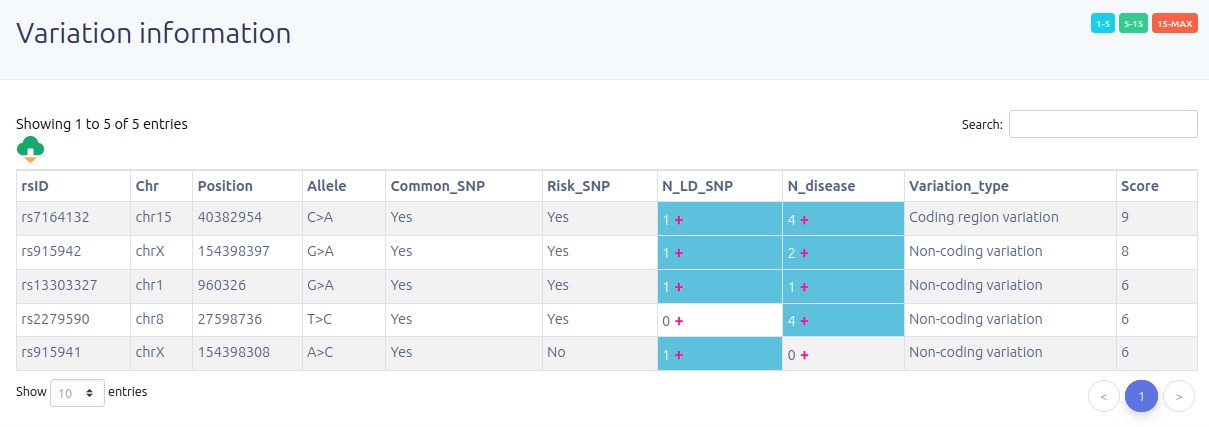
eRNAs that intersected with variants.
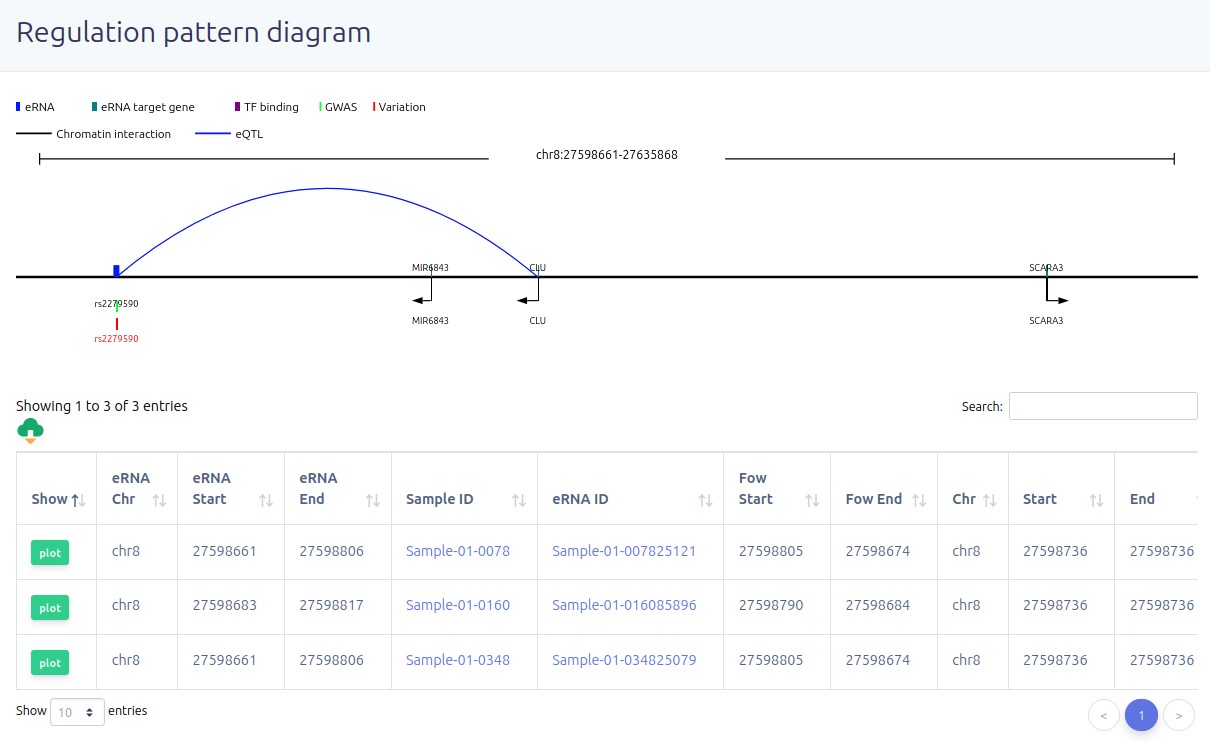
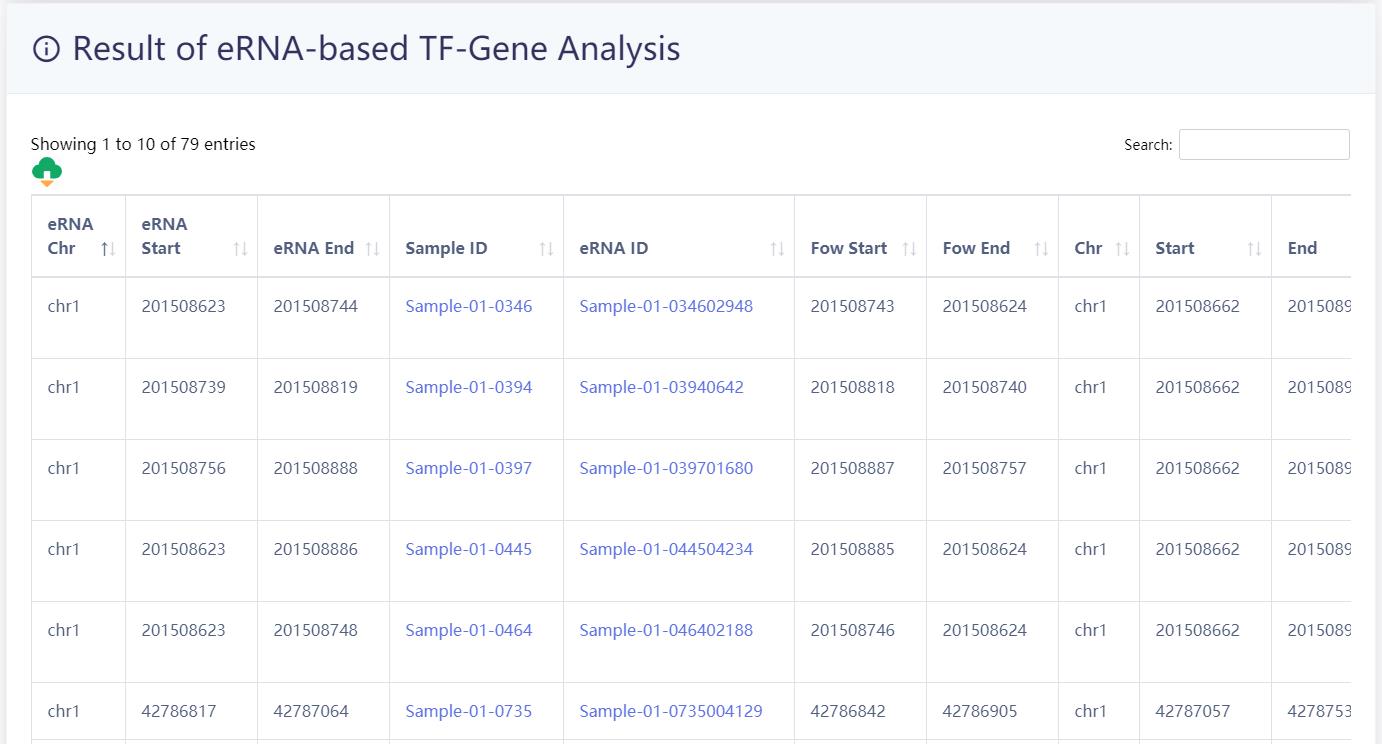
URL:https://bio.liclab.net/eRNAbase/analysis/eRNA_TF_gene.php
4.3.3 eRNA-mediated TF-target gene analysis
Users submit two lists of TF and gene and select species of interest, eRNAbase identifies eRNA-mediated TF-gene pair(s) in different tissue types. Meanwhile, the users can set different TF identification methods through the 'Type' option.
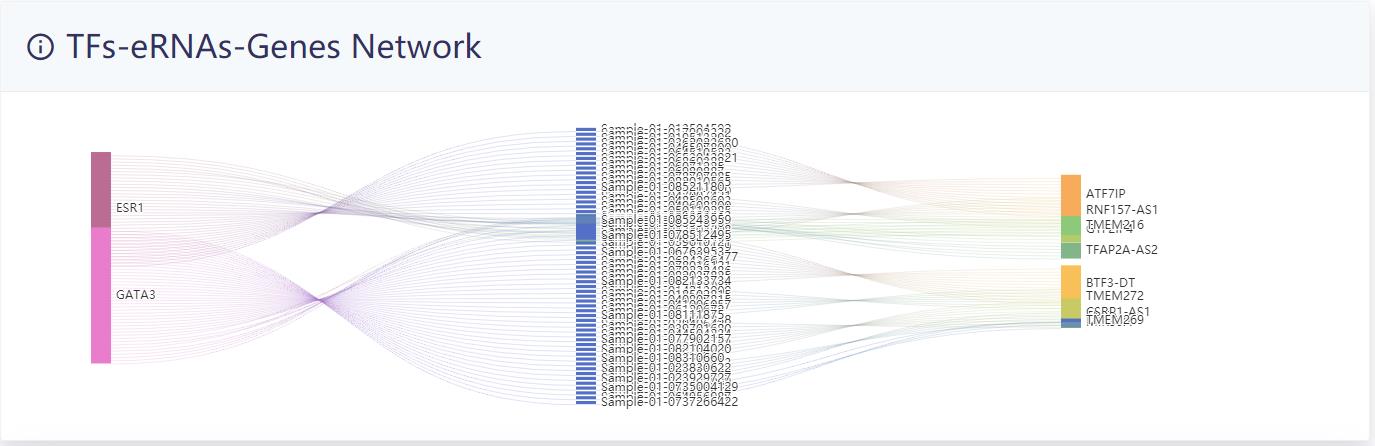
The results of the analysis of the example data are as follows.
TF-eRNA-gene network
URL:https://bio.liclab.net/eRNAbase/download.php
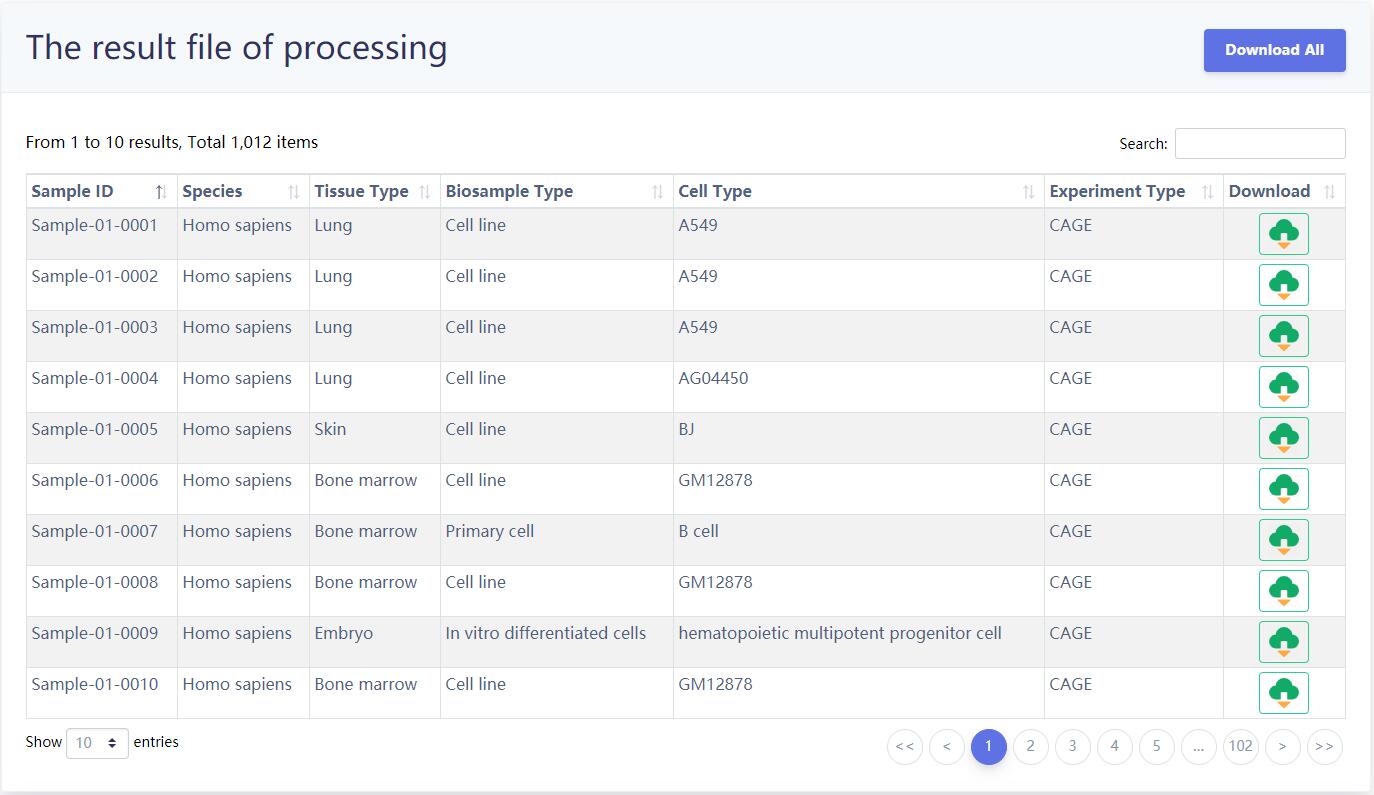
4.5 Download
The results of the eRNA information for each sample as well as the index table of the difference samples are provided for download in the 'Download' page. Users can quickly download associated information. In addition, we also provided the batch download and non-redundant eRNA sets download functions for users.
5. Development environment
The current version of eRNAbase was developed using MySQL 5.7.17 (http://www.mysql.com) and runs on a Linux-based Apache Web server (http://www.apache.org). We used PHP 7.0 (http://www.php.net) for server-side scripting. We designed and built the interactive interface using Bootstrap v3.3.7 (https://v3.bootcss.com) and JQuery v2.1.1 (http://jquery. com). We used ECharts (http://echarts.baidu.com) and D3 (https://d3js.org) as a graphical visualization framework, and JBrowse (http://jbrowse.org) is the genome browser framework. We recommend using a modern web browser that supports the HTML5 standard such as Firefox, Google Chrome, Safari, Opera or IE 9.0+ for the best display.
The eRNAbase database is freely available to the research community using the web link (https://bio.liclab.net/eRNAbase/index.php). Users are not required to register or login to access features in the database.
6. Material disclaimer
The materials and frameworks used by eRNAbase are shared by the network and do not contain intellectual property infringement. If there is any infringement, please write to us and we will change it in time.