Help&FAQ
- What information is available in the AgingReG?
- What information is included in the AgingReG?
- Database content and construction
- How to use the AgingReG?
- Explanation of the definitions used by the website
- Frequently Asked Questions
- Development environment
Aging factors, including genes, compounds, or external factors, such as radiation, hypoxia and lipid-lowering therapy, are very important for studying the mechanism of aging and the treatment of aging related diseases. In fact, the regulation of gene expression during aging plays a crucial role in specific cells and tissues.
We established a manually curated database of aging factors (AgingReG, https://bio.liclab.net/Aging-ReG/index.php ), focusing on the regulatory relationships during aging with experimental evidence in human. The database features include aging genes information (i.e. target genes, accelerate / prevent aging, regulatory relationships); their mediated annotation (i.e. risk SNPs and eQTLs); external factors and pathways that related to aging; functional annotation of aging gene related pathways; identification of aging genes by sequence comparison. We expect that AgingReG will help users thoroughly understand the biological processes and in-depth regulatory mechanisms of aging process.
Through reviewing 1784 articles that have been screened, the current release of AgingReG documents 1345 aging gene entries (involving 321 upregulating pairs, 363 downregulating pairs, 160 activation-regulating pairs and 32 Inhibition-regulating pairs), 804 external factor entries (involving 330 upregulating pairs, 269 downregulating pairs, 170 activation-regulating pairs and 2 Inhibition-regulating pairs) and 8 aging-related pathway entries. Especially, unlike existing aging related gene databases, AgingReG focuses on the regulatory relationships between aging factors and target genes confirmed by experiments, and visualizes them.
By searching the hallmarks of aging as keywords, AgingReG retrieved the most comprehensive aging related articles to the greatest possible extent and extracts information according to rigorous standards. AgingReG provides two categories of the detailed information of aging factors: basic information and regulatory information.
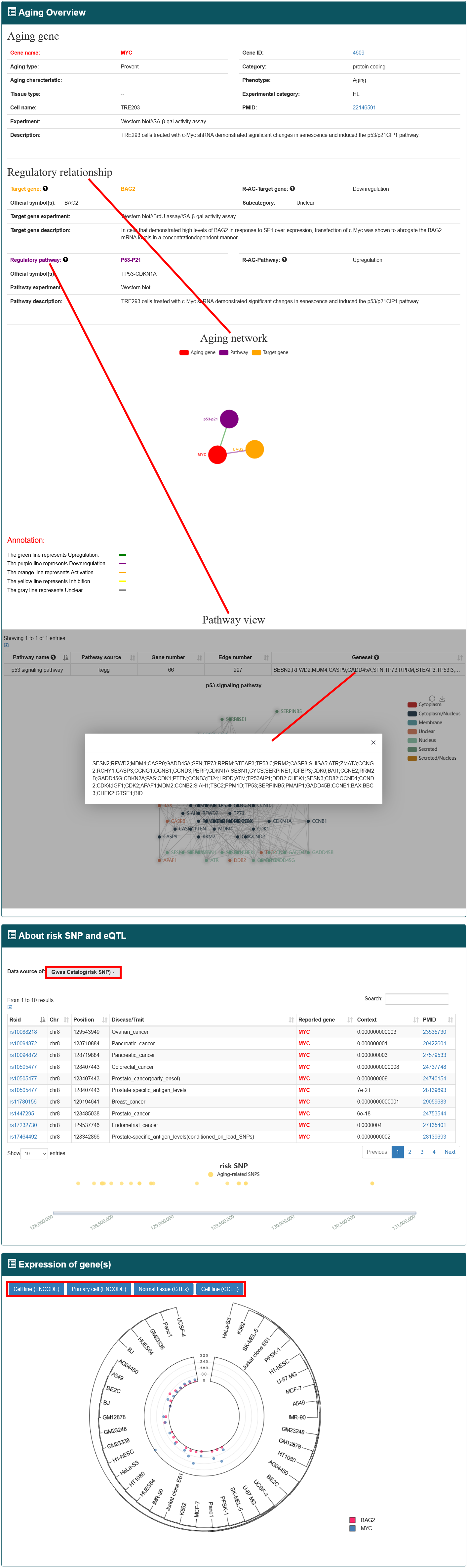
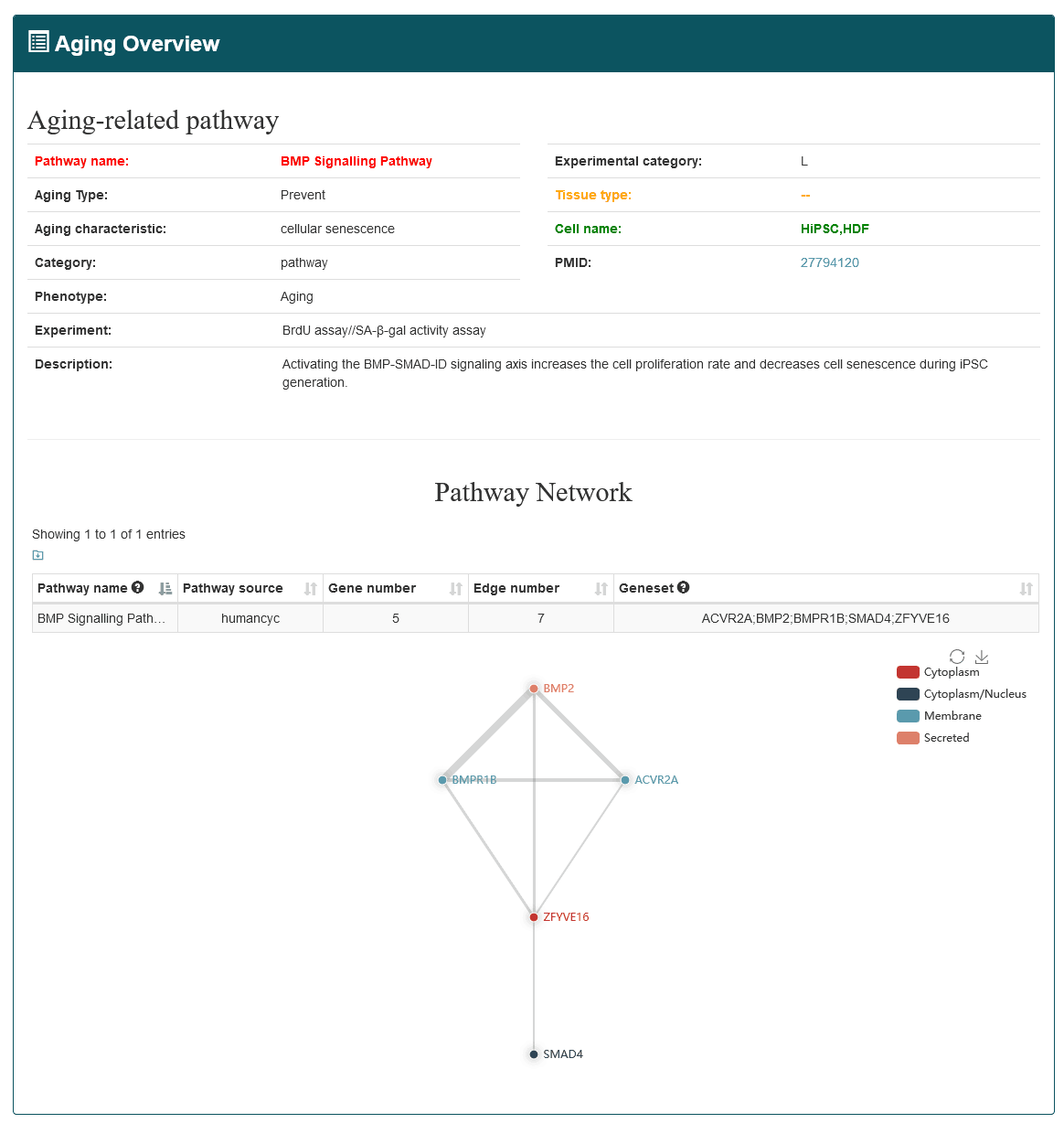
'Basic information' includes gene name, gene ID, category (protein coding or ncRNA), aging type (accelerate or prevent), aging characteristic (the hallmarks of aging), cell name, tissue type, experimental technology (such as SA-β-gal activity assay, western blot, knockdown, and luciferase report assay), experiment category (low-throughput or high-throughput) and the experimental descriptions recorded in the literature. AgingReG also includes genetic and epigenetic annotations for each aging gene, including common risk single-nucleotide polymorphisms (SNPs) and expression quantitative trait loci (eQTLs).
'Regulatory information' includes target genes and regulatory pathways, whose expression is regulated and changed during aging. In the regulatory relationship category, the regulatory information of aging factors includes information on the targets' name, regulatory type (upregulation, downregulation, activation, Inhibition and unclear), experiments and description. The relationships between aging genes and their regulatory objects were named 'R-AG-Target gene' (relationships between aging gene and target gene) and 'R-AG-Pathway' (relationships between aging gene and pathway).Based on the regulatory relationships, AgingReg also provides some subcategories about modifications based on the regulatory relationships, including “binding to promoter”, “phosphorylation”, “dimerization”, “acetylation”, “ubiquitylation” and “unclear”.
Users can search aging factors through four paths, including 'Search by Gene name' (select regulatory type and input gene name of interest), 'Search by External factors' (select regulatory type and input external factors of interest), 'Search by Pathway' (select regulatory type and pathway name of interest), 'Search by Tissue type and Cell name' (select aging factor type and input tissue type and cell name). All the information of aging factors can be obtained by entering the 'Start search'.

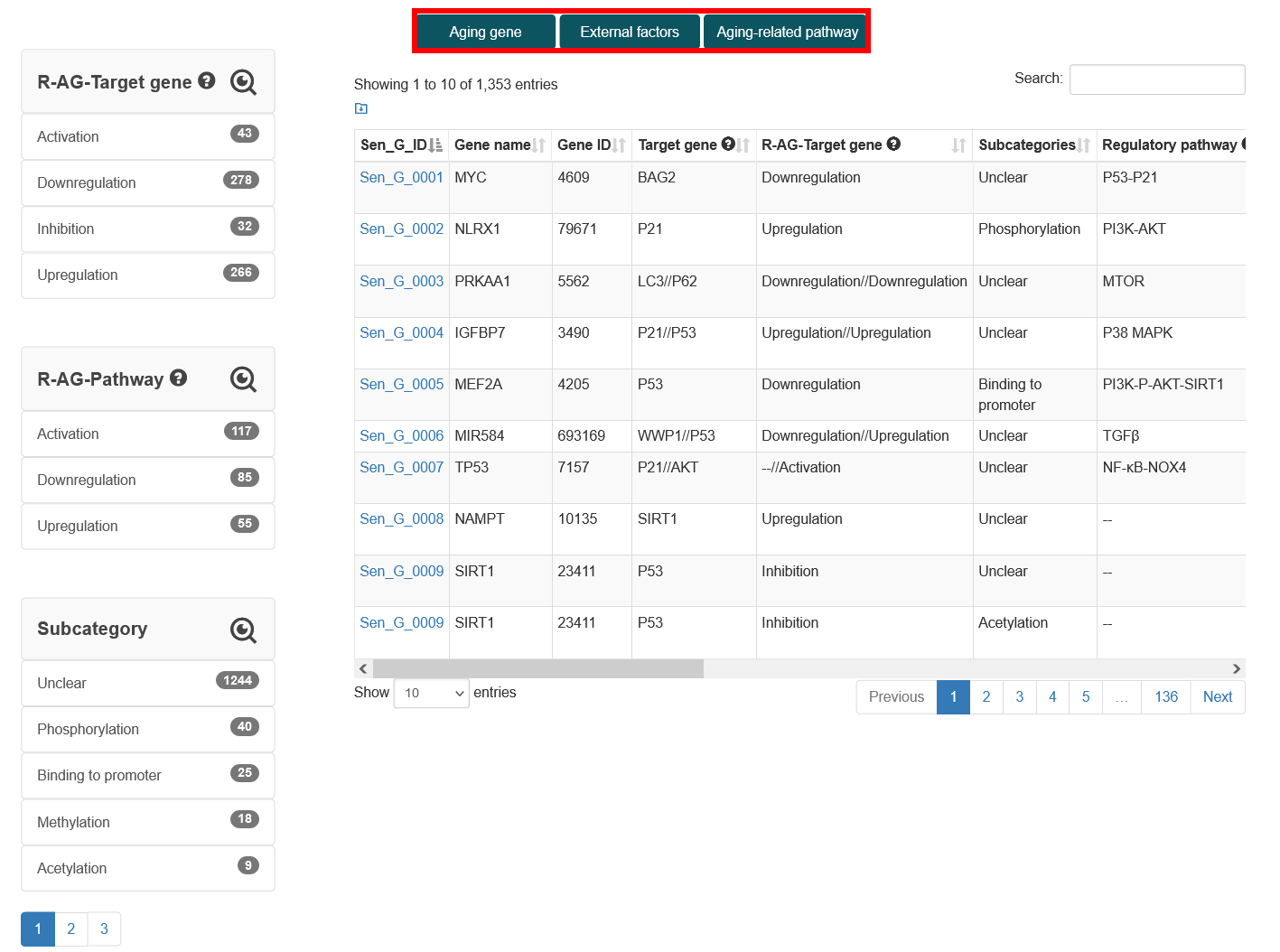
The 'Data-Browse' page is organized as an interactive alphanumeric sorting table, allowing users to quickly browse 'Gene name', 'Gene ID', 'Target gene', 'Regulatory pathway', 'Phenotype' and 'Aging type'. Users can quickly browse the regulatory relationships, aging genes, and related information of interest through the fuzzy search function and easily change the number of records per page through the 'Display item' drop-down menu.
4.2. Search
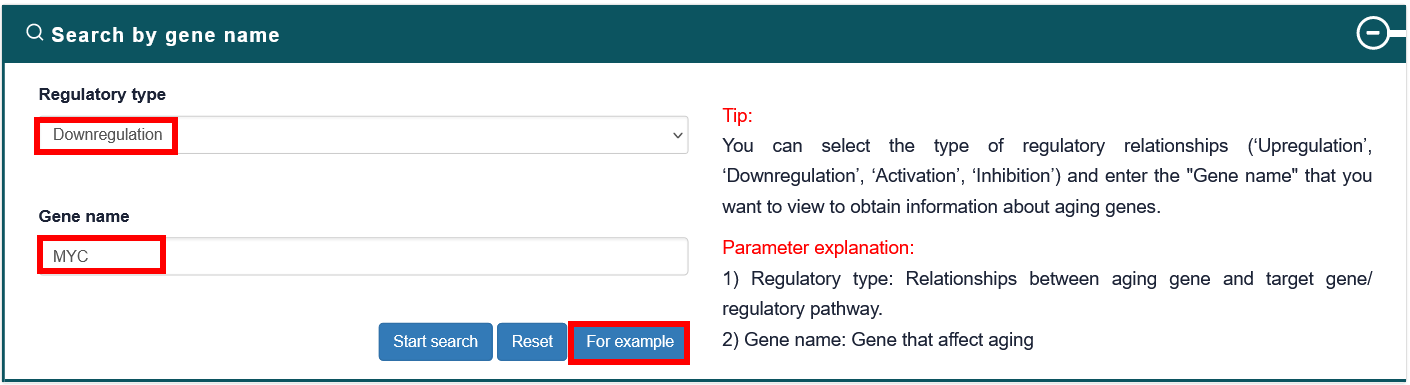
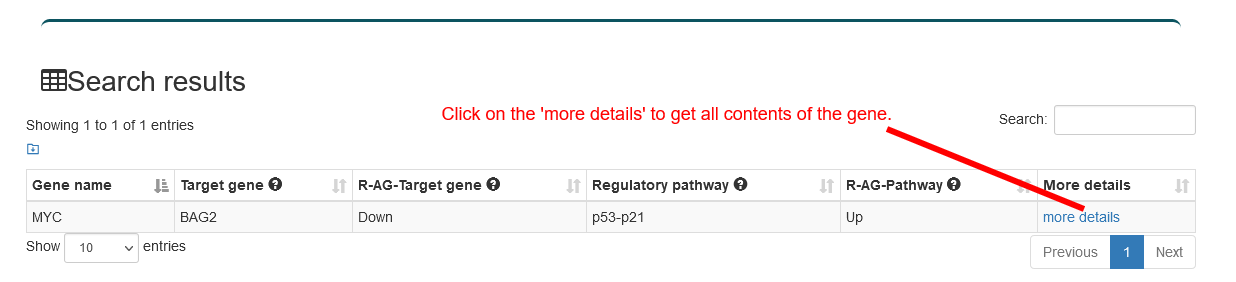
Users can select the type of regulatory relationship and input a gene name of interest, the search results will be displayed on the next page. Users can click the 'More Details' to view the details.

4.2.1. Search by Gene Name
Users can select the type of regulatory relationship and input a gene name of interest, the search results will be displayed on the next page. Users can click the 'More Details' to view the details.
4.2.2. Search by Category and External factors
Users can select the type of regulatory relationship and input an external factor name of interest, the search results will be displayed on the next page. Users can click the 'More Details' to view the details.
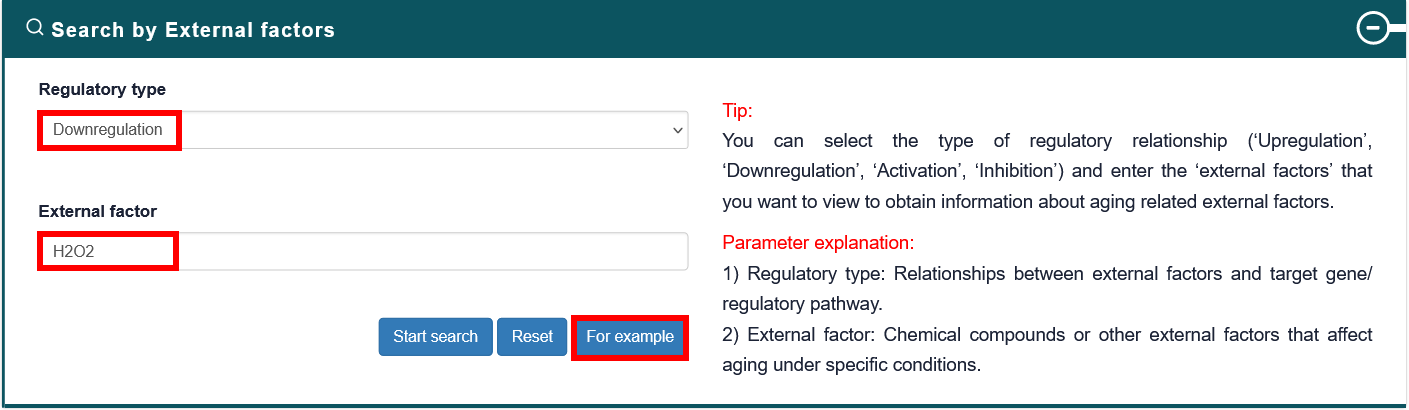
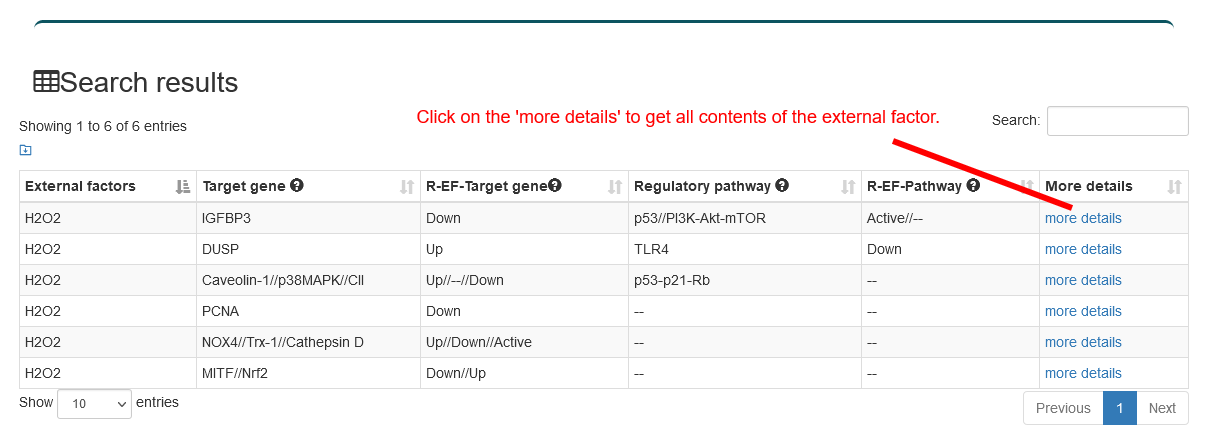
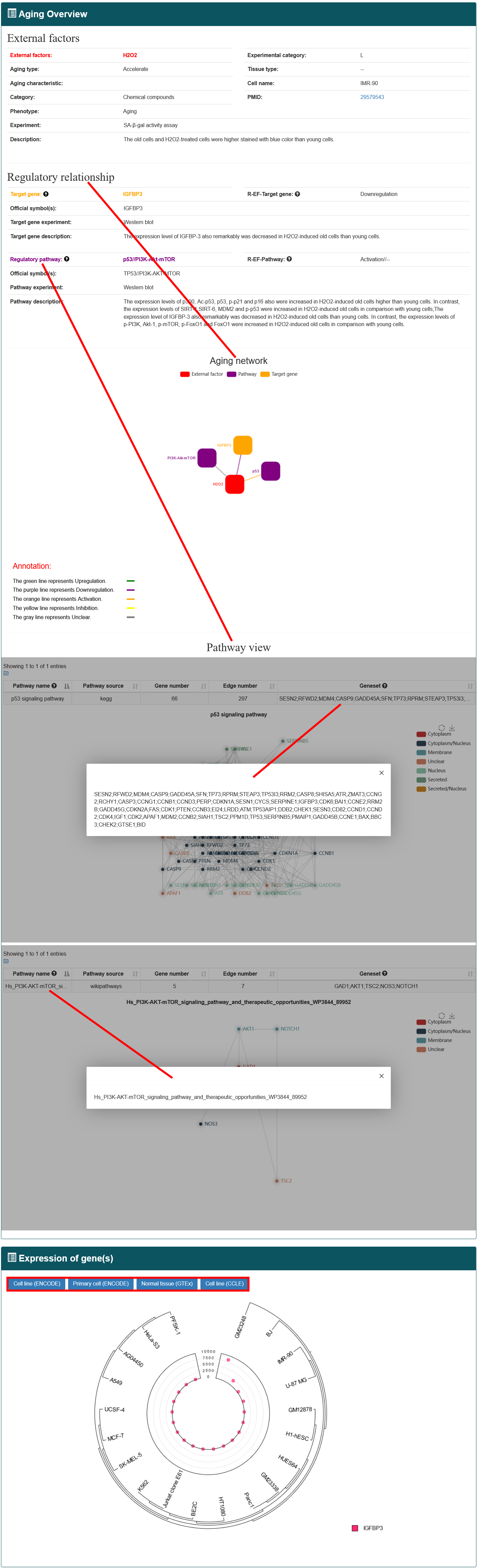
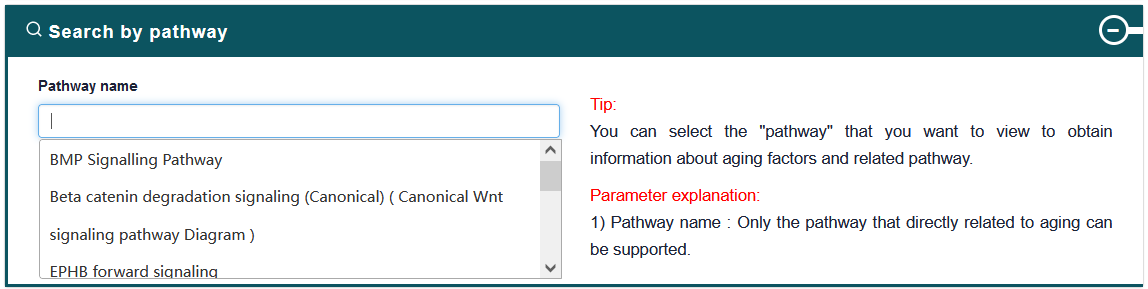
4.2.3. Search by Pathway
Users can input a pathway name of interest, the search results will be displayed on the next page. Users can click the 'More Details' to view the details.
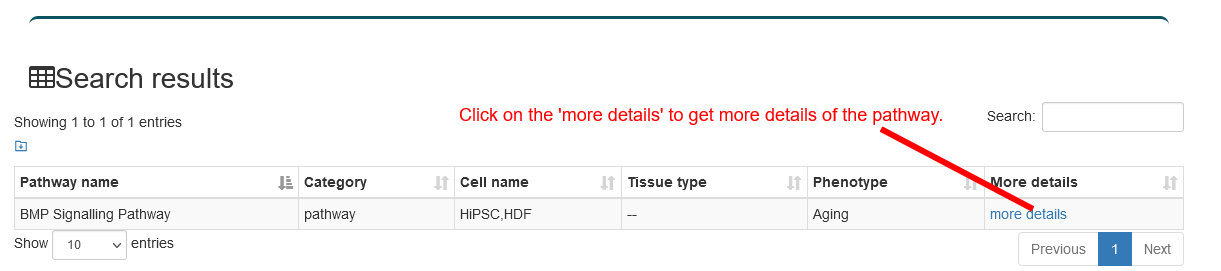
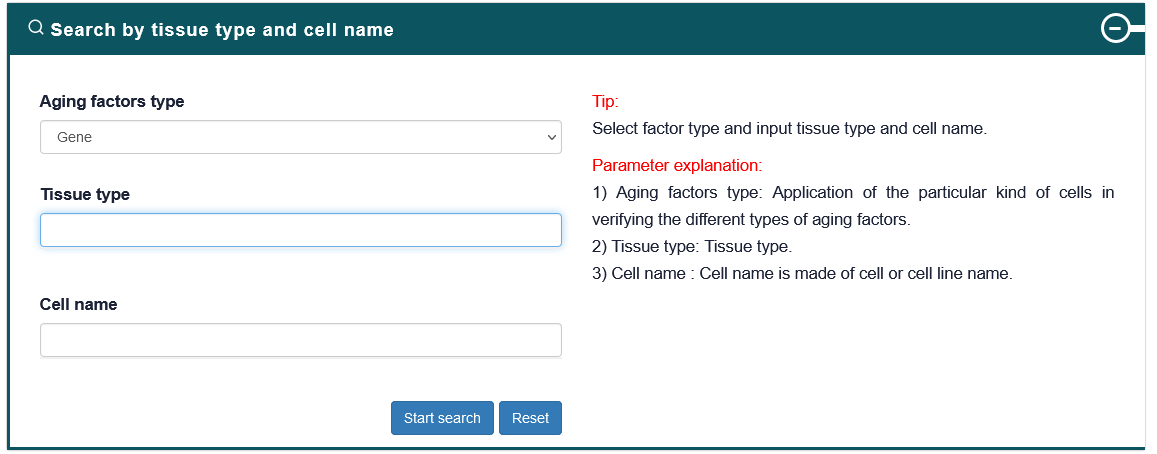
4.2.4. Search by Tissue type and Cell name
Users can choose the aging factor type and input tissue type and cell name of interest, the search results will be displayed on the next page. Users can click the 'More Details' to view the details.
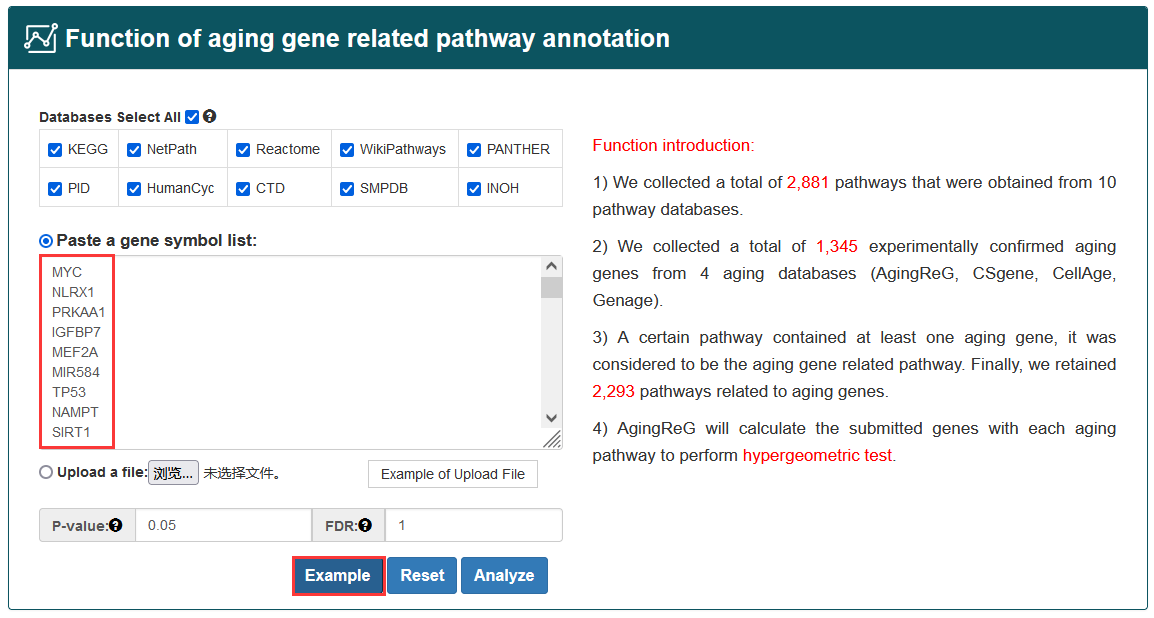
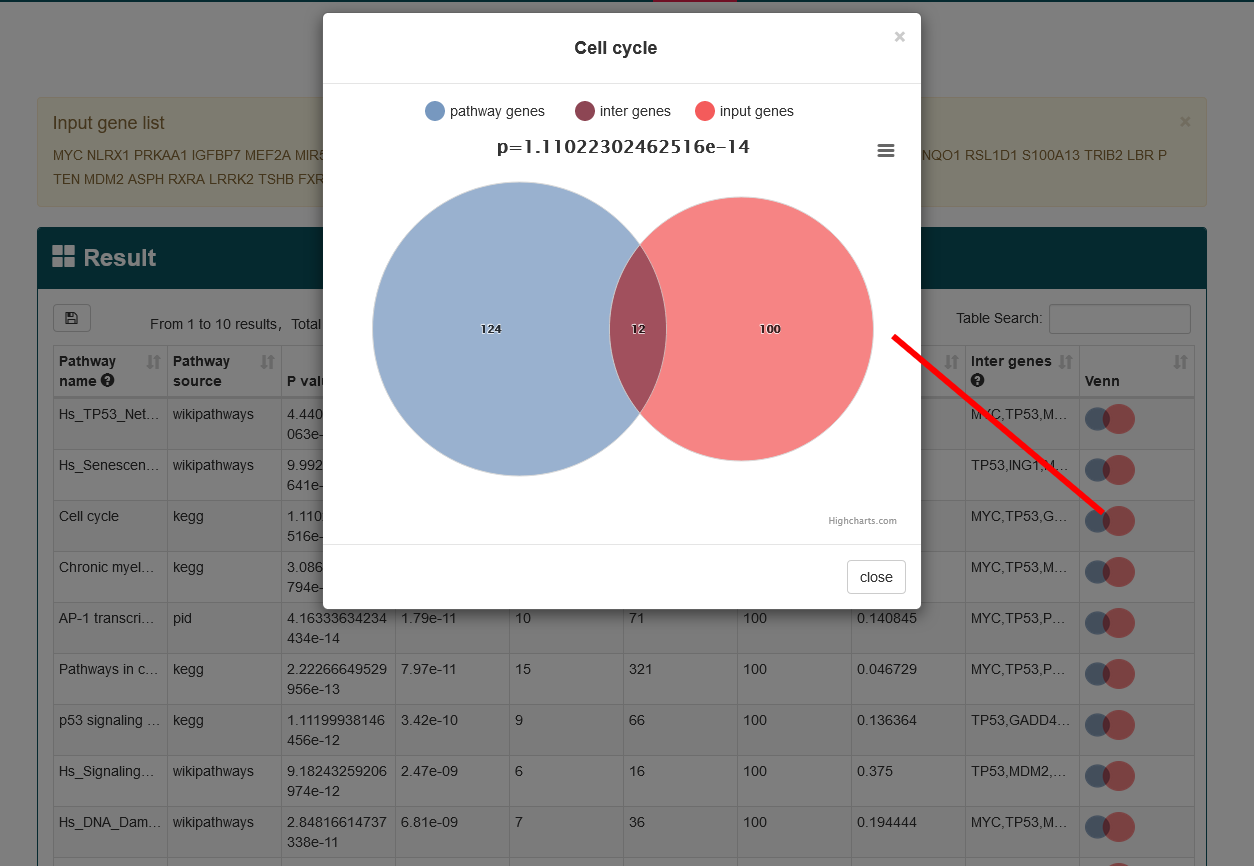
To better understand the biological significance behind aging genes, AgingReG provides researchers with a comprehensive functional annotation of aging pathway. In AgingReG, user can manually input a set of genes of interest (one gene symbol per line) or upload the gene set in a text file and then select at least one pathway database (containing KEGG, Reactome, NetPath, WikiPathways, PANTHER, PID, HumanCyc, CTD, SMPDB, and INOH) to perform function of aging pathway annotation. User can also modify the number of genes inputted and set the p value to control the stringency of the analysis.
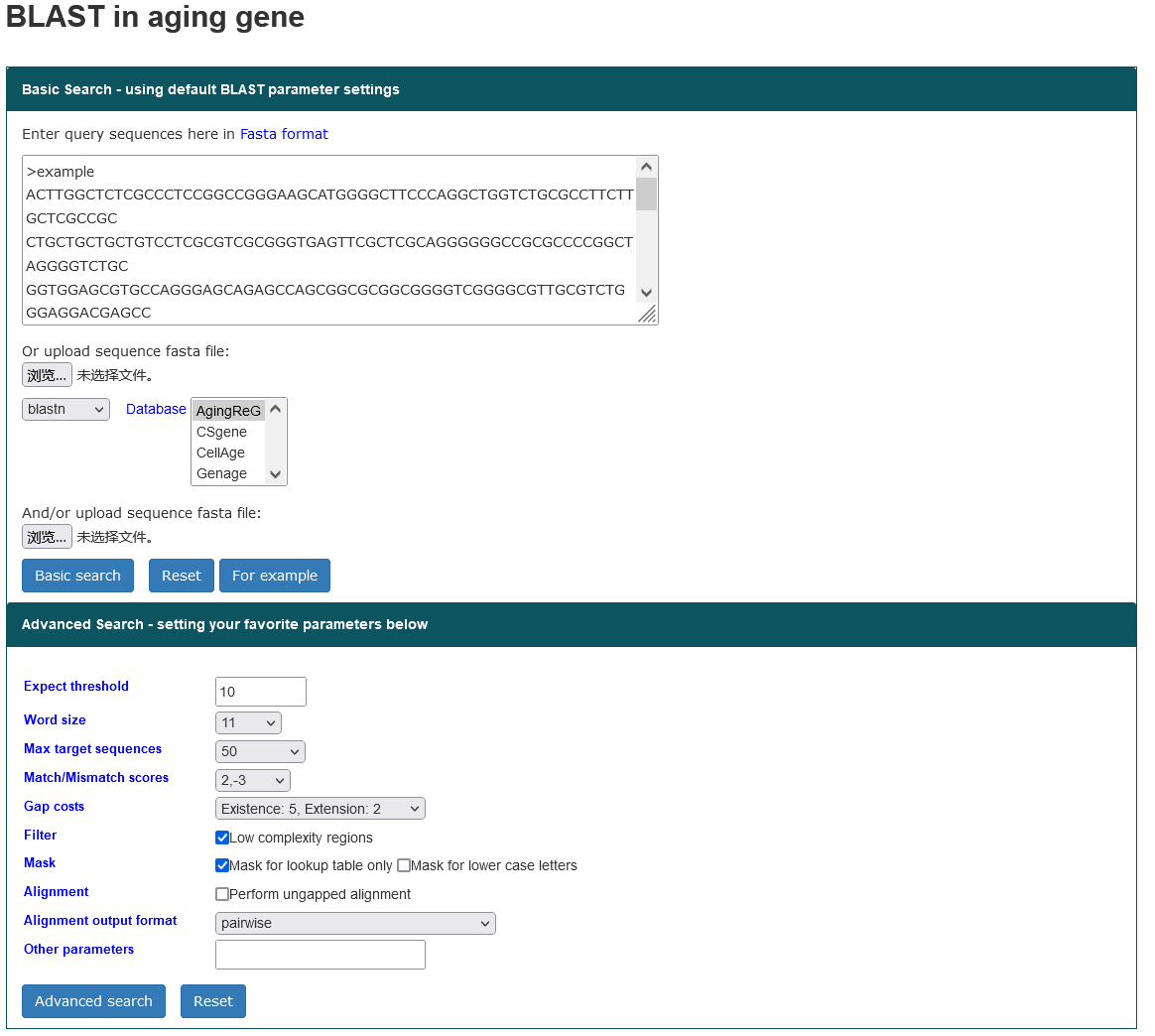
4.4.1. Tool-BLAST
The AgingReG database provides practical tool for comparing nucleotide and amino acid sequences. To help users identify potential aging related genes, we performed a multigene sequence comparison. The sequences of each gene or protein of interest were aligned against the other experimentally confirmed aging related genes by BLAST, and the similarity of the sequences was obtained and provided for users' reference.
Steps to start a BLAST search:
- Copy and paste multiple query sequences in fasta format to text box, or upload a sequence file in FASTA format.
- Select one database to blast against or upload user's own sequence dataset file in FASTA format.
- Select search features to get more specific information (optional).
- Click 'Basic search' button to start search.
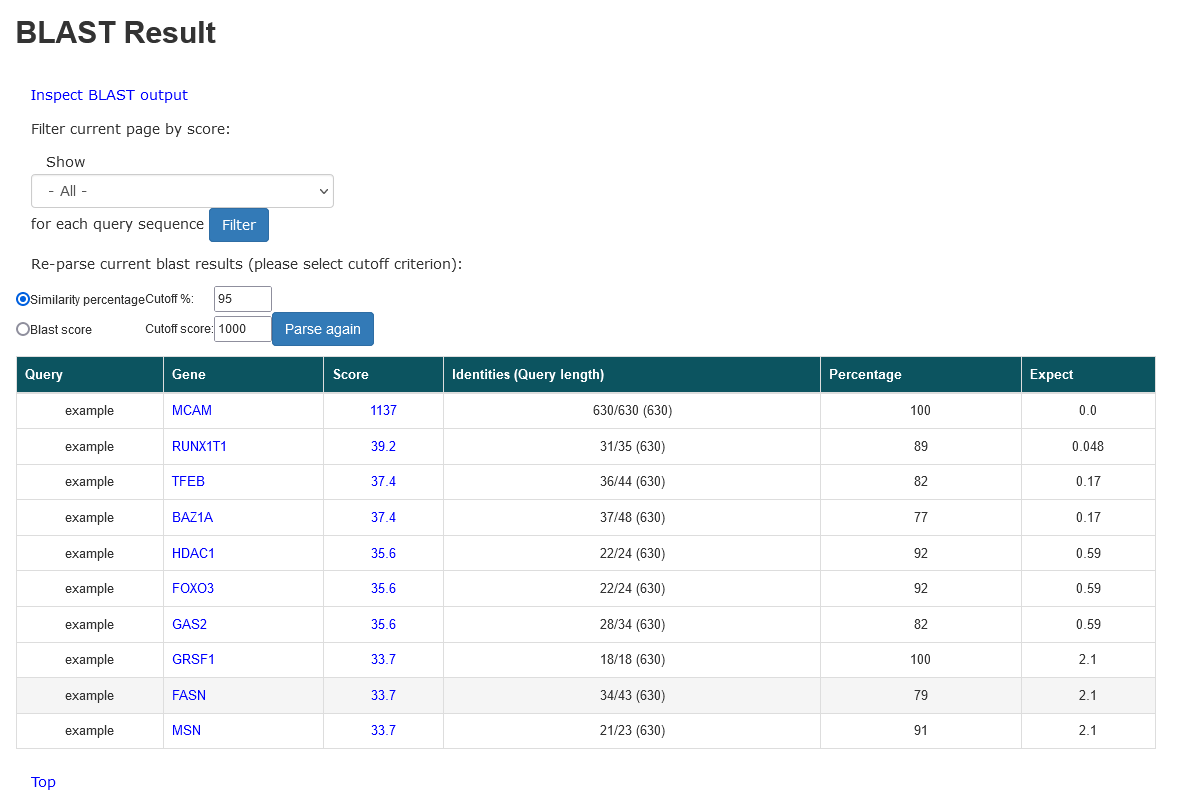
Friendly output to easily parse and navigate the BLAST result.
- User can reset different parameters to filter and parse again the BLAST result.
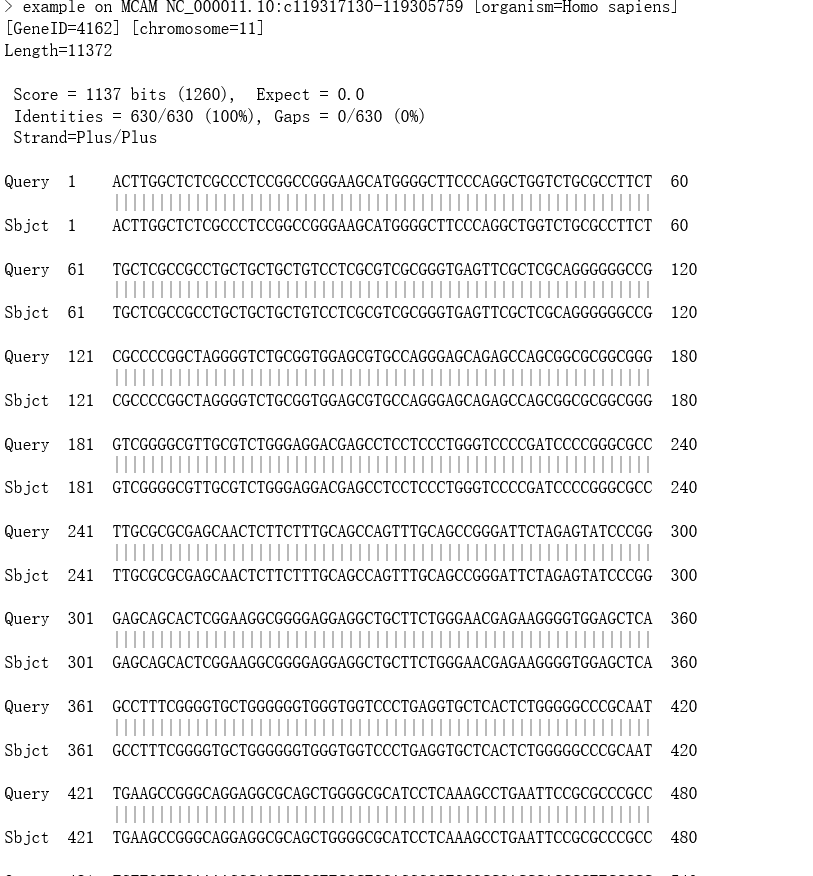
- Click one of links in Score field will locate the pair-wise alignment between the query sequence and subject sequence in the blast result file.
- Click "Inspect BLAST output" link will open the blast result file. The BLAST automatically divides a big output file into several small files.

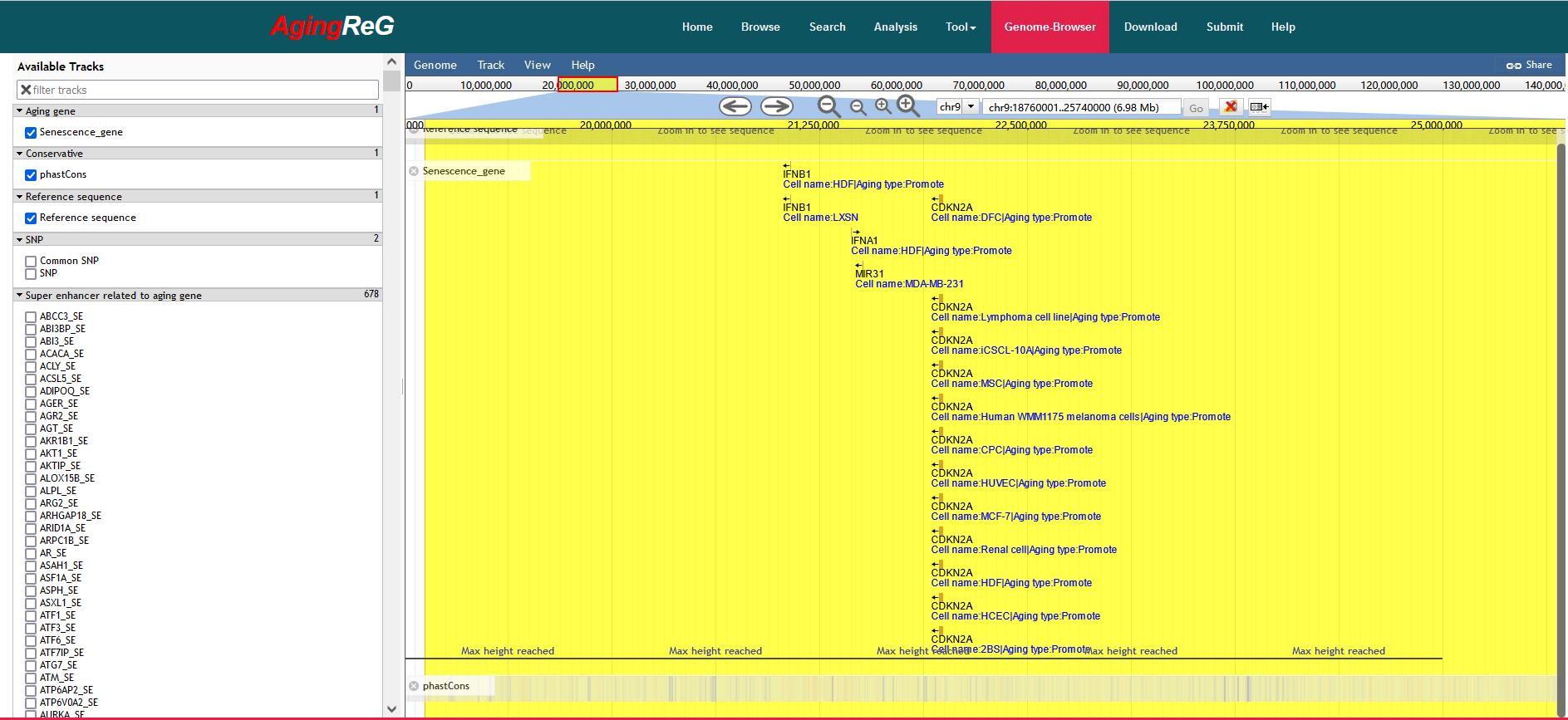
To help users view the proximity of aging genes and their mediation annotation in the genome, we developed a personalized genome browser using JBrowse and added many useful tracks, such as aging genes and related risk SNPs. In addition, the roles played by aging genes in different studies and the types of cells analyzed can be simultaneously displayed.
The data of aging factors of all entries are provided for download in the 'Download' page. Users can download their interested files.
| AgingReG contains 20 columns separated by tab: | Description: |
|---|---|
| PMID | The PMID of the article. |
| Gene name | The name of aging-related gene. |
| Gene ID | The gene ID of the aging-related gene. |
| Category | Category of aging-related gene,including protein coding or ncRNA. |
| Cell name | The specific cells used in the aging-related experiments. |
| Tissue type | The specific tissues used in the aging-related experiments. |
| Phenotype | The name of disease or pathological process. |
| Aging type | Accelerate or prevent aging. |
| Experiment | The name of experiment used to confirm the effect of gene on aging. |
| Description | Experimental description of the effect of genes on aging. |
| Target gene | The target gene of aging-related gene confirmed by experiments. |
| Target gene experiment | The name of experiment used to confirm the target gene. |
| Target gene description | Experimental description of how aging genes regulate targets. |
| R-AG-Target gene | Relationships between aging gene and target gene. |
| Regulatory pathway | The regulatory pathway of aging-related gene confirmed by experiments. |
| Pathway experiment | The name of experiment used to confirm the pathway. |
| Pathway description | Experimental description of how aging genes regulate pathways. |
| R-AG-Pathway | Relationships between aging gene and pathway. |
| Experimental category | The experimental category(High throughput+Low throughput or just Low throughput experiments). |
| Aging characteristic | The hallmarks of aging. |
| AgingReG contains 19 columns separated by tab: | Description: |
|---|---|
| PMID | The PMID of the article. |
| External factors | The name of the external factors that have an effect on aging. |
| Category | Chemical compounds or others. |
| Cell name | The specific cells used in the aging-related experiments. |
| Tissue type | The specific tissues used in the aging-related experiments. |
| Phenotype | The name of disease or pathological process. |
| Aging type | Accelerate or prevent aging. |
| Experiment | The experiments used to confirm the effect on aging. |
| Description | Experimental description of the effect of external factors on aging. |
| Target gene | The target gene confirmed by experiments. |
| Target gene experiment | The name of experiment used to confirm the target gene. |
| Target gene description | Experimental description of the regulation on target gene. |
| R-EF-Target gene | Relationships between external factors and target gene. |
| Regulatory pathway | The regulatory pathway of external factors confirmed by experiments. |
| Pathway experiment | The name of experiment used to confirm the pathway. |
| Pathway description | Experimental description of how external factors regulate pathways. |
| R-EF-Pathway | Relationships between external factors and pathway. |
| Experimental category | The experimental category(High throughput+Low throughput or just Low throughput experiments). |
| Aging characteristic | The hallmarks of aging. |
| AgingReG contains 11 columns separated by tab: | Description: |
|---|---|
| PMID | The PMID of the article. |
| Pathway name | The name of the pathway that have an effect on aging. |
| Category | Pathway. |
| Cell name | The specific cells used in the aging-related experiments. |
| Tissue type | The specific tissues used in the aging-related experiments. |
| Phenotype | The name of disease or pathological process. |
| Aging type | Accelerate or prevent aging. |
| Experiment | The name of experiment used to confirm the effect of the pathway on aging. |
| Description | Experimental description of the effect of pathway on aging. |
| Experimental category | The experimental category(High throughput+Low throughput or just Low throughput experiments). |
| Aging characteristic | The hallmarks of aging. |
6.1. Why is some information fruitless?
Reply: It may be because there is no corresponding annotation for the gene or the region has no result.
6.2. Why might web pages load slowly?
Reply: AgingReG has advanced storage technology and sufficient bandwidth to meet the needs of most users for the speed of web page loading. However, it is not excluded that few users have poor user experience due to network reasons.
6.3.What's the meaning of the symbol '//' and '-'?
Reply:When an aging gene regulates several target genes, we use the symbol ‘//’ to distinguish and correspond to each regulatory relationship. 'A//B' indicates that two target genes 'A' and 'B' are respectively regulated by aging factors. For example, in 'PRKAA1' entry, target gene 'SOD2//SIRT3' and the regulatory type of target gene 'Up//Up' indicate that PRKAA1 influences aging by upregulating the expression of SOD2 and SIRT3 respectively. 'A-B' in the regulatory pathway represents upstream protein A and downstream protein B in the same pathway.
Development environment
In the recent version of AgingReG, the graphical visualization box is based on ECharts and Highcharts, and it was built using JBrowse as the Genome browser framework. The design and construction of the interactive interface were based on Bootstrap v3.3.7 and JQuery v2.1.1. The workflow of the database was developed using MySQL 5.7.17, and the server-side script was PHP 7.0 running on a Linux-based Apache Web server. To obtain the best display effect, we recommend using a modern web browser that supports the HTML5 standard, such as Firefox, Google Chrome, Safari, Opera, or ie9.0+.
In addition, for ease of use, users can access the database without registering or logging in.