AgingReGV1.0
A curated database of Aging Regulatory Relationships in human
What is AgingReG?
Aging and cellular senescence are characterized by progressive loss of physiological integrity, which is correlated with the pathogenesis of serious diseases, including cancer, cardiac and neural diseases. Aging and cellular senescence can be induced by physiological, pathological, and external factors (such as radiation, hypoxia and lipid-lowering therapy). Numerous studies show that gene regulatory events play crucial roles in the aging process.
Hence, we established a manually curated database of aging factors (AgingReG, https://bio.liclab.net/Aging-ReG/ ), focusing on the regulatory relationships during aging with experimental evidence in human. By curating thousands of published literature, 2157 aging factors entries (1345 aging genes entries, 804 external factors entries and 8 aging related pathway entries) and related regulatory information were manually curated, and the regulatory relationships were classified into four types according to their functions: 1) Upregulation, which indicates that aging factors up-regulate the expression of target genes during aging. 2) Downregulation, which indicates that aging factors down-regulate the expression of target genes during aging. 3) Activation, which indicates that aging factors enhance the activity of target genes during aging. 4) Inhibition, which indicates that aging factors inhibit the activation of target molecule activity, lead to target 's activity declined or lost. . AgingReG involves 651 upregulating pairs, 632 downregulating pairs, 330 activation-regulating pairs and 34 Inhibition-regulating pairs, covering 195 disease types and more than 800 kinds of cells and tissues from 1784 published literature. AgingReG provides a user-friendly interface to query, browse and visualize the detailed information about the regulatory relationships during aging.
How can I use AgingReG?
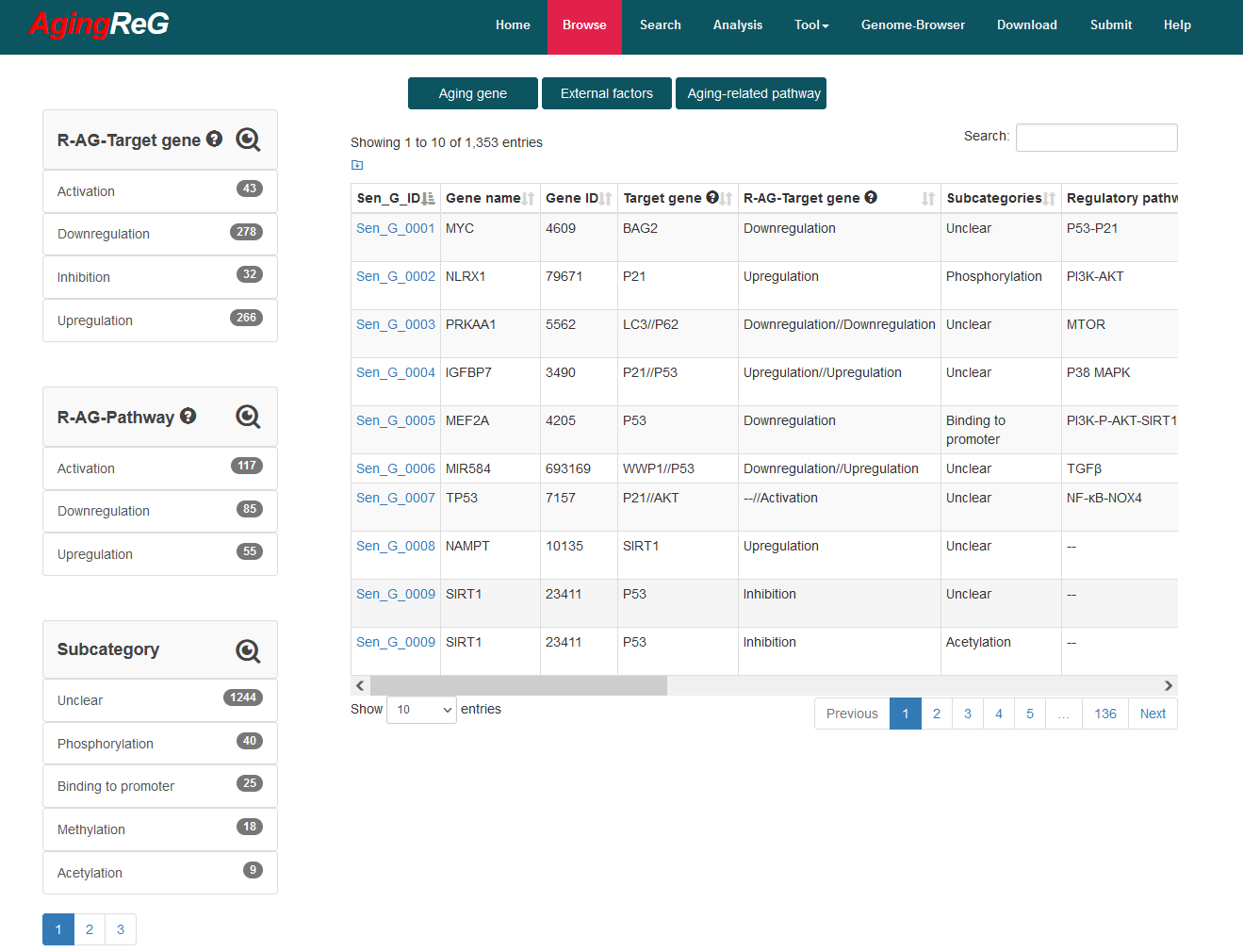
Browse aging related information: The ‘Data-Browse’ page is organized as an interactive and alphanumerically sortable table, allowing users to quickly browse ‘Gene name’, ‘Gene ID’, ‘Target gene’, ‘R-AG-Target gene’/ ‘R-EF-Target gene’, ‘Regulatory pathway’, ‘R-AG-pathway’/ ‘R-EF-pathway’, ‘Phenotype’ and ‘Aging type’. The ‘Show entries’ drop-down menu is provided for changing the number of records per page conveniently.
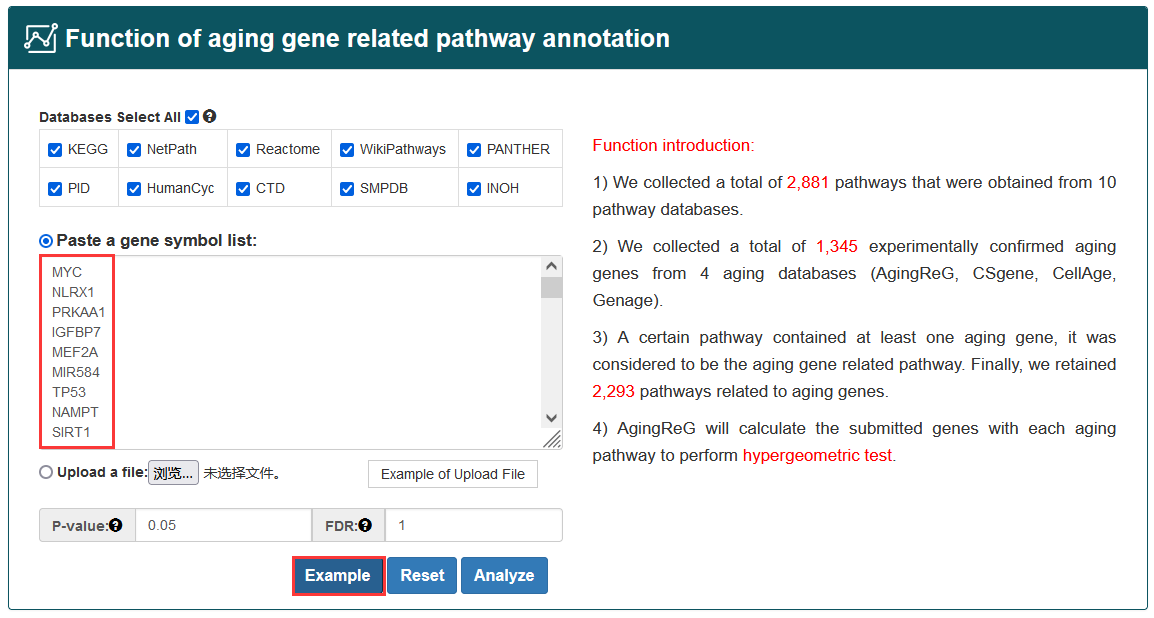
Query aging factors: Users can search aging factors through four paths, including ‘Search by Gene name’ (select regulatory type and input gene name of interest), ‘Search by External factors’ (select regulatory type and input external factors of interest), ‘Search by Pathway’ (select regulatory type and pathway name of interest), ‘Search by Tissue type and Cell name’ (select aging factor type and input tissue type and cell name). All the information of aging factors can be obtained by entering the ‘Start search’.
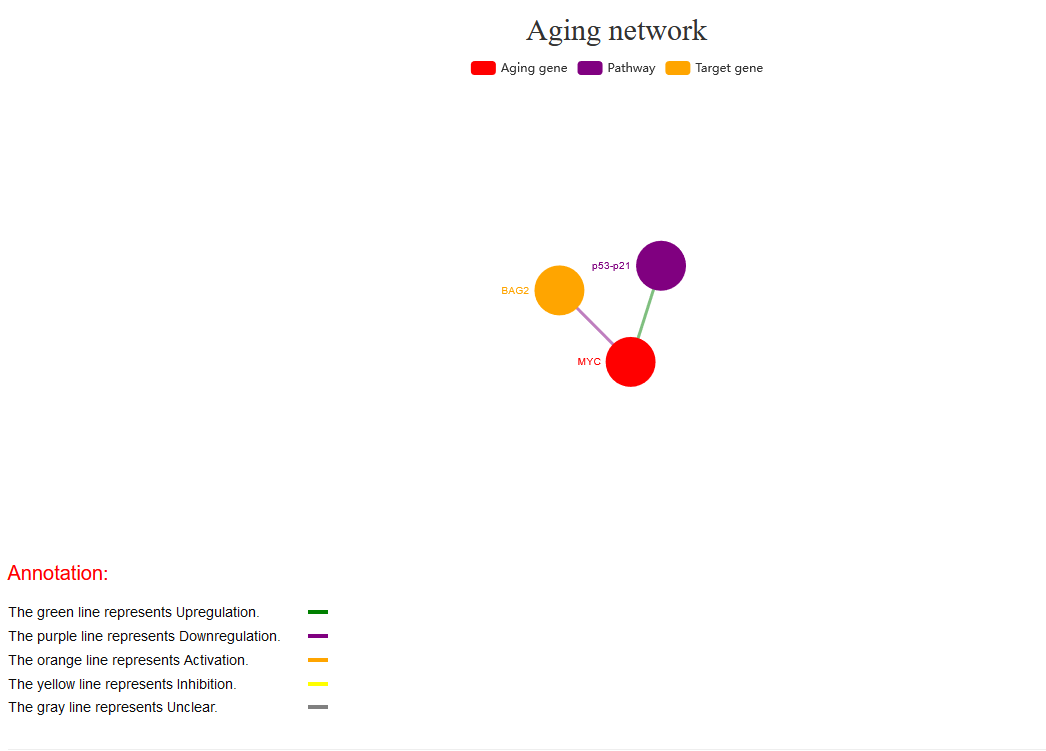
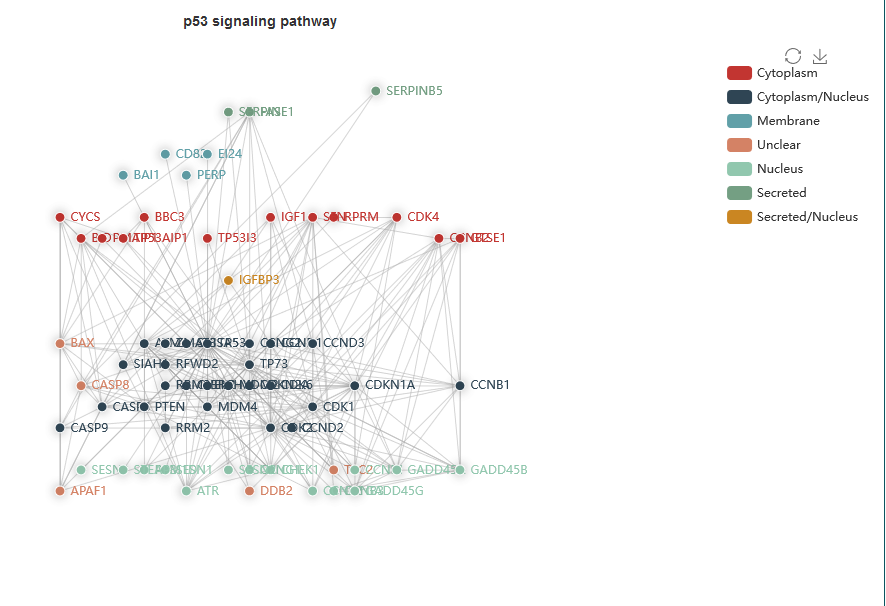
Aging factors details: Users can click ‘More details’ to view the detailed information of each aging factor, such as the relationship between aging genes, including basal gene annotation (‘Aging gene name’, ‘Target gene’, ‘Gene ID’, ‘Category’ and ‘Regulatory pathway’), literature records (‘Phenotype’, ‘Aging type’, ‘Aging characteristic’, ‘Tissue type’, and ‘Experiment category’) and descriptions of experiments in the literature. In addition, the network is visually displayed, showing the relationships between aging genes and their regulatory information, with different coloured lines representing regulatory relationships. Furthermore, the pathway network involved in aging genes and the expression of target genes in cells is visualized.