Introduction
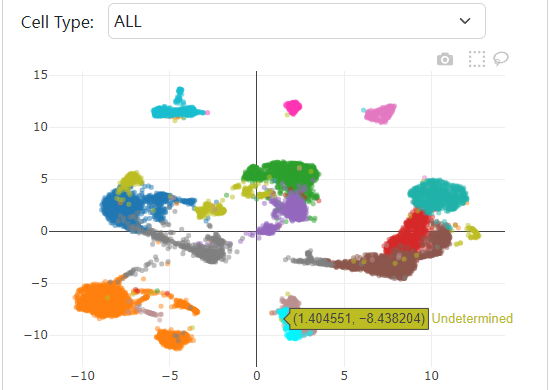
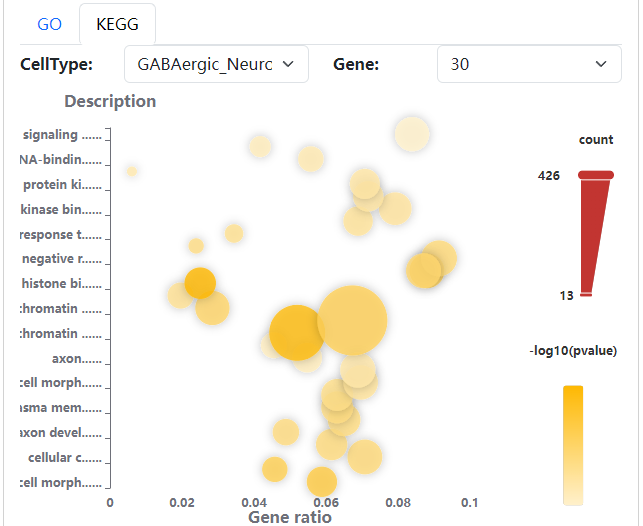
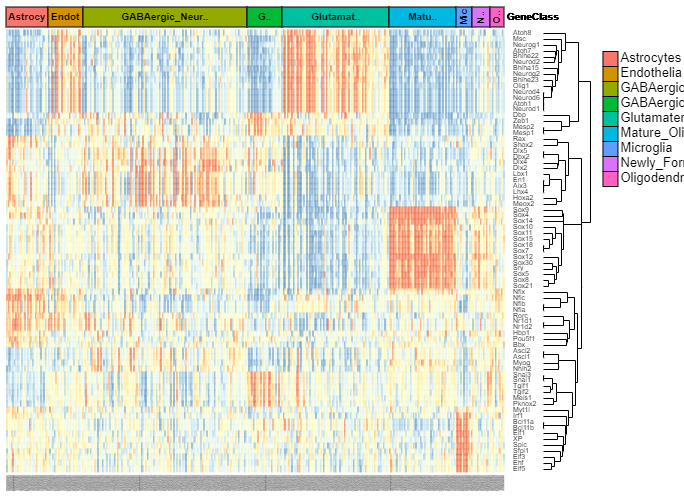
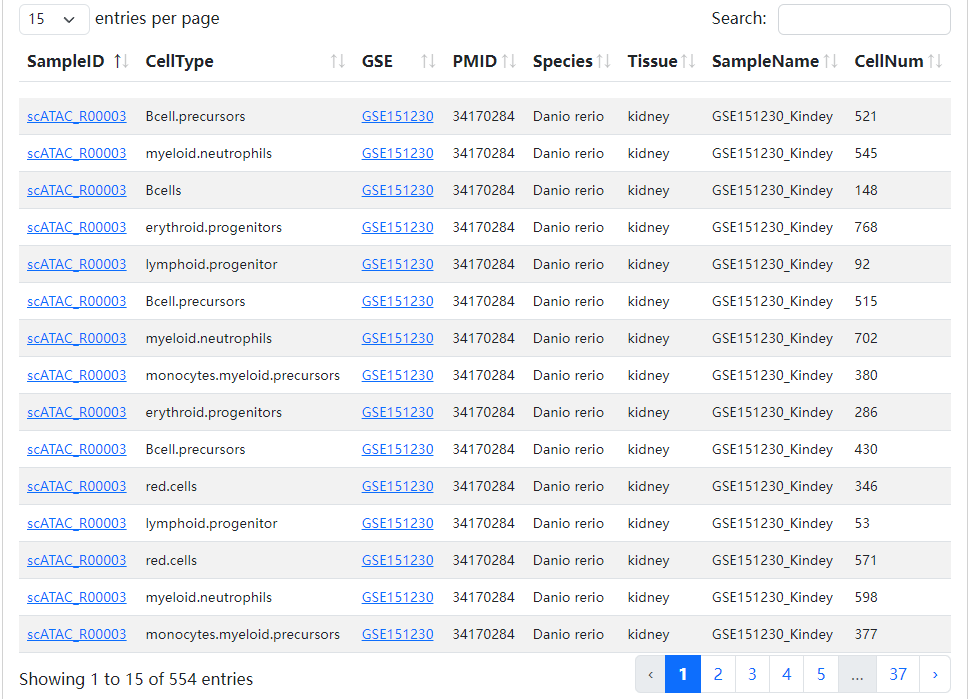
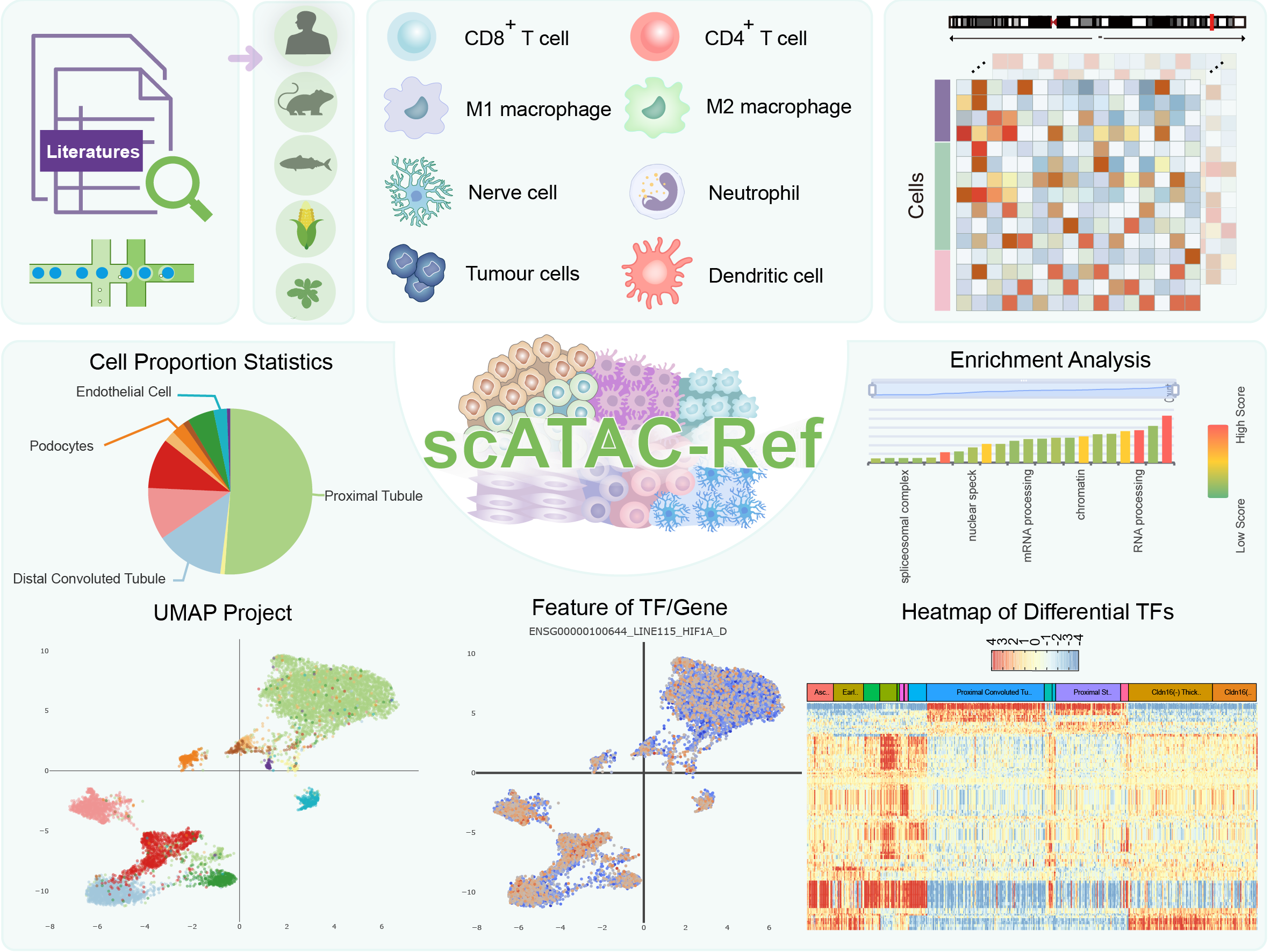
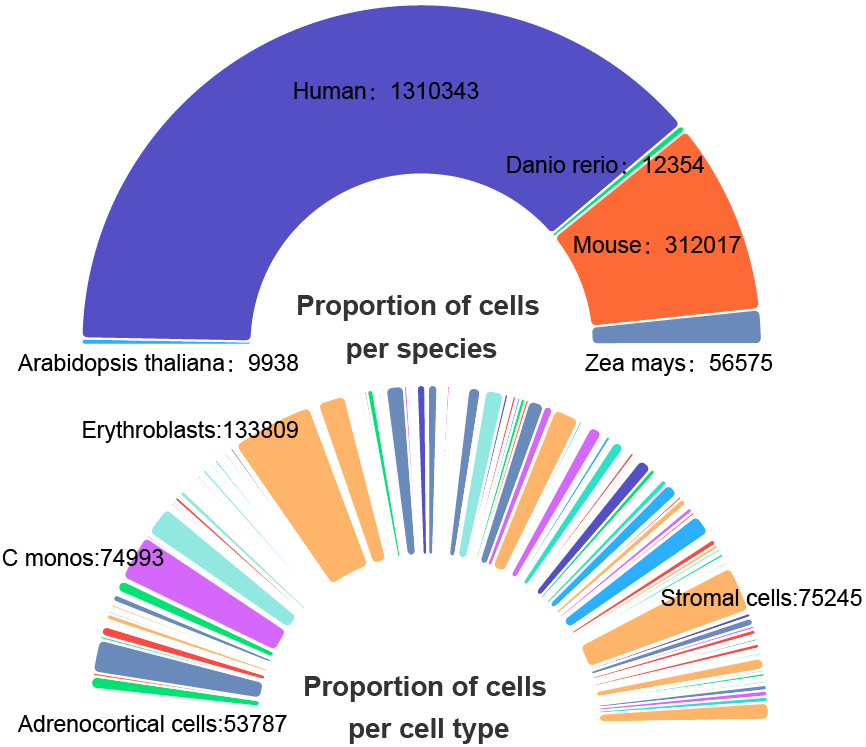
We developed a database, scATAC-Ref (https://bio.liclab.net/scATAC-Ref/), to provide a large-scale resource of scATAC-seq data with known cell labels in five species. The current version of scATAC-Ref documents > 400 cell/tissue types manually annotated by published articles, covering 1,694,372 cells across five species: Human, Mouse, Danio rerio, Zea mays and Arabidopsis thaliana. Furthermore, comprehensive genome-wide chromatin accessibility analysis for known cells/tissues is crucial for exploring cell/tissue heterogeneity, and can help uncover key cell subtype and potential markers. Hence, for these scATAC-seq data with known cell labels, we used a uniform system environment and software parameters to calculate gene activity score, TF enrichment score, differential accessibility region (DAR) and pathway/GO term enrichment. The scATAC-Ref also provided a convenient, user-friendly interface to query, browse and visualize cell types of interest, to help elucidate cell type related functions and potential biological effects.
Statistic
Sister Projects
|
The comprehensive human Super-Enhancer database |
|
|
A web tool for super-enhancer associated regulatory analysis |
|
|
An experimentally supported enhancer database for human and mouse |
|
|
A comprehensive human chromatin accessibility database |
|
|
A comprehensive database of human transcriptional regulation of lncRNAs |
|
|
A resource for transcriptional regulation information of circRNAs |
|
|
A comprehensive human gene expression profile database with knockdown/knockout of transcription factors |
|
|
A variation annotation database for human |
|
|
A comprehensive human lncRNA sets resource and enrichment analysis platform |
Live Statistics
For publication of results please cite the following article
Cite: Qian FC, Zhou LW, Zhu YB, Li YY, Yu ZM, Feng CC, Fang QL, Zhao Y, Cai FH, Wang QY, Tang HF, Li CQ. scATAC-Ref: a reference of scATAC-seq with known cell labels in multiple species. Nucleic Acids Res. 2024 Jan 5;52(D1):D285-D292. doi: 10.1093/nar/gkad924.