What is LncSEA 2.0?
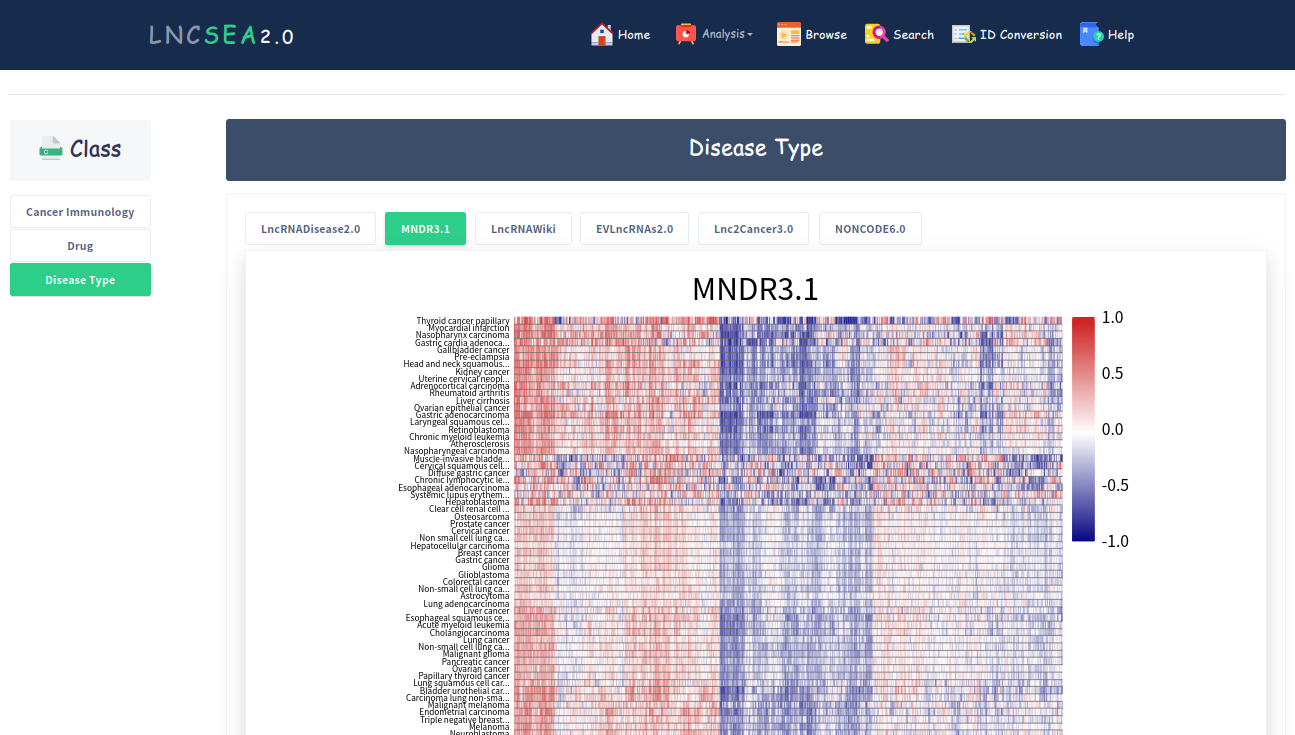
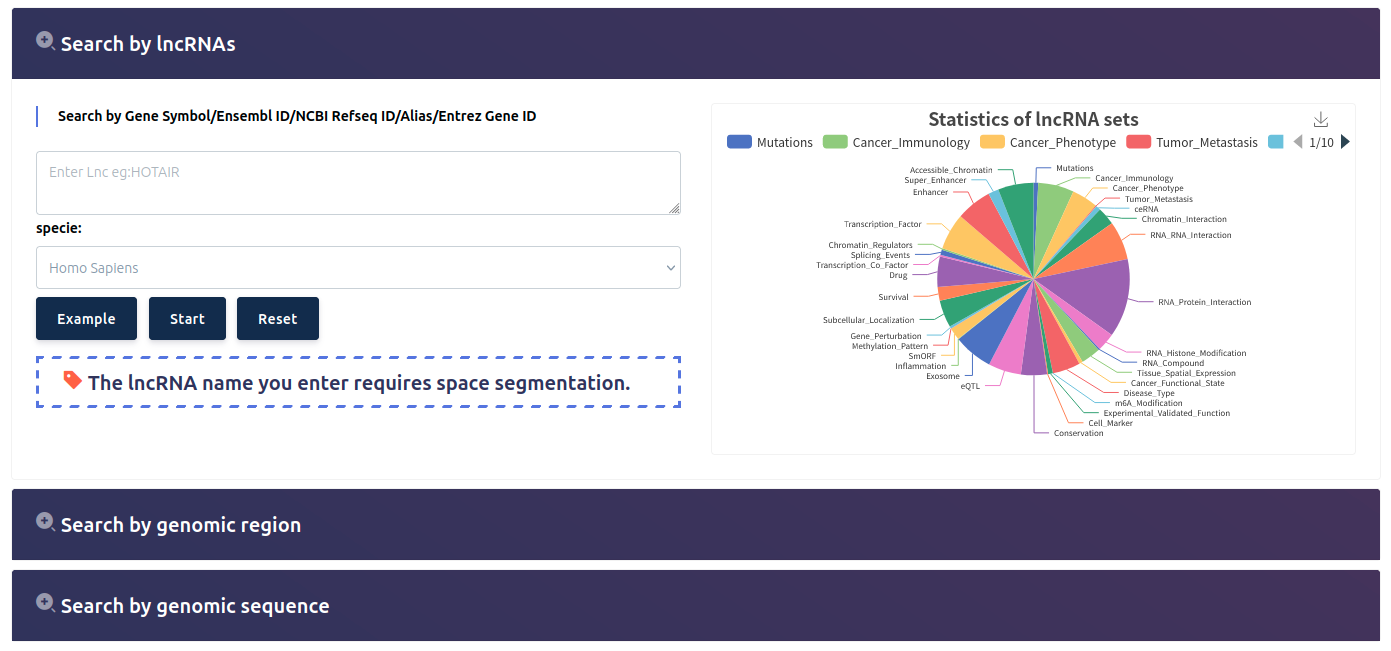
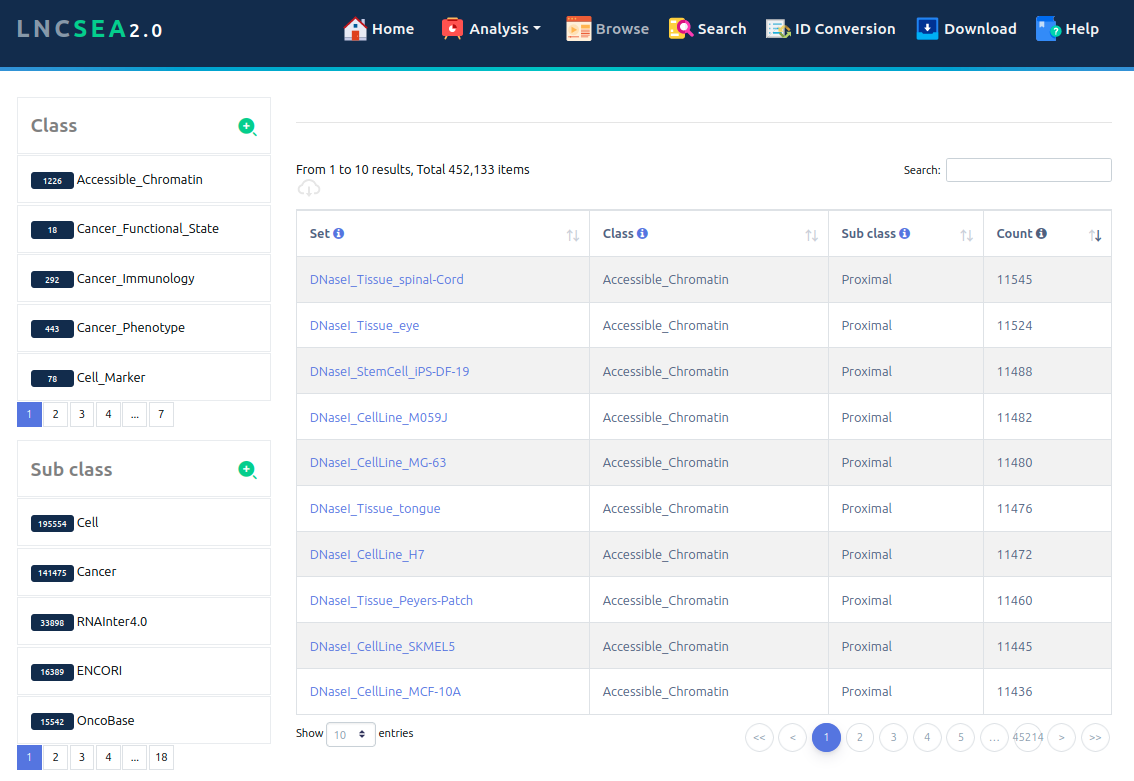
LncSEA 2.0 supports more than 400,000 reference lncRNA sets involved in 33 categories (Mutations, Cancer Immunology, Tumor Metastasis, ceRNA, Chromatin Interaction, RNA-RNA Interaction, RNA-Protein Interaction, RNA Histone Modification, RNA Compound, Tissue Spatial Expression, Cancer Functional State, Disease Type, m6A Modification, Experimental Validated Function, Cell Marker, Conservation, eQTL, Exosome, Inflammation, SmORF, Methylation Pattern, Gene Perturbation, Subcellular Localization, Survival, Drug, Transcription Co Factor, Splicing Events, Chromatin Regulators) and 86 sub-categories, which covered over 200,000 lncRNAs. We not only collected lncRNA sets based on downstream regulatory data sources but also calculated a large number of lncRNA sets regulated by upstream transcription factors (TFs) and DNA regulatory elements by integrating TF ChIP-seq, DNase-seq, ATAC-seq and H3K27ac ChIP-seq data associated with hundreds of human cell types.
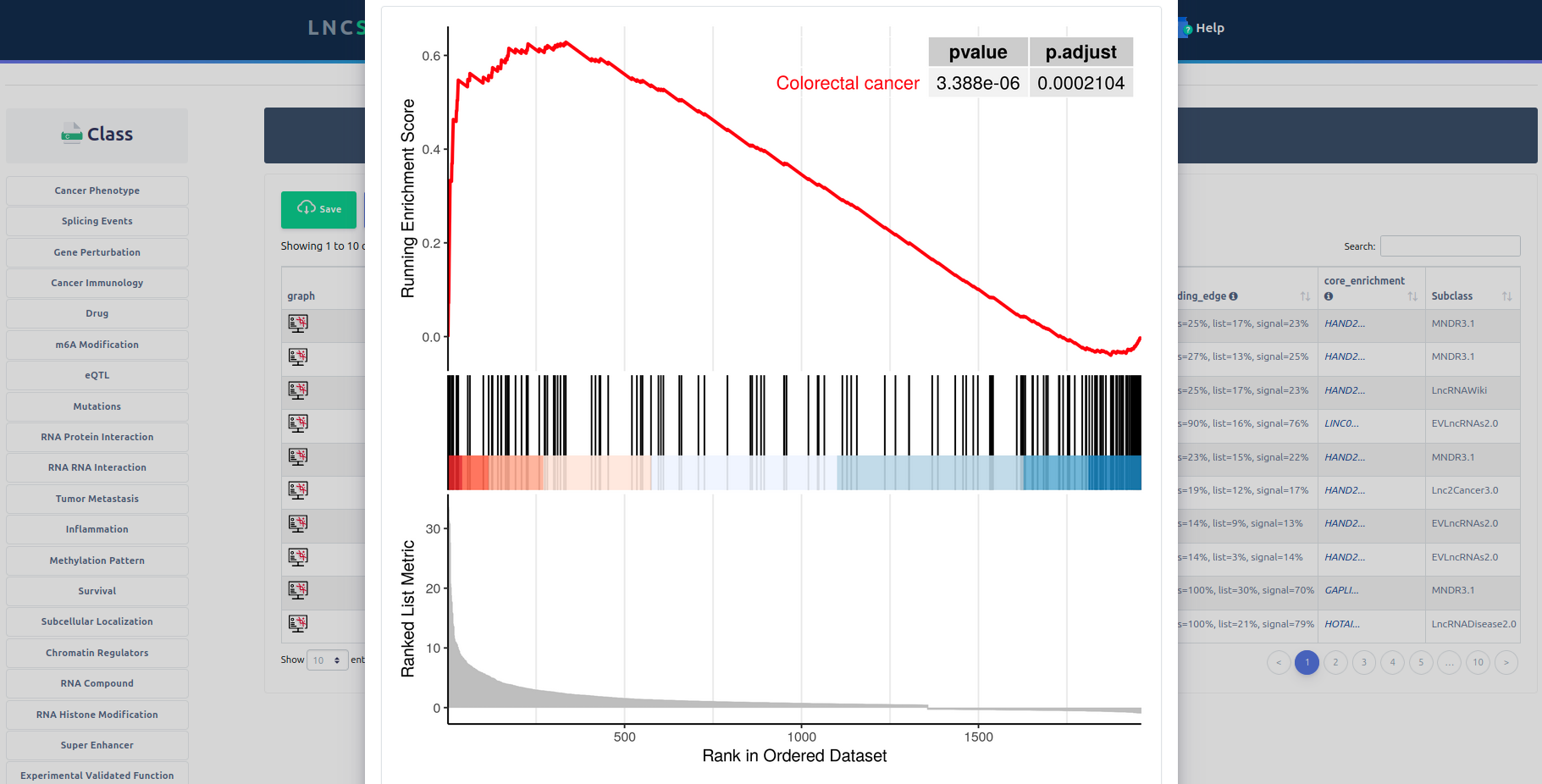
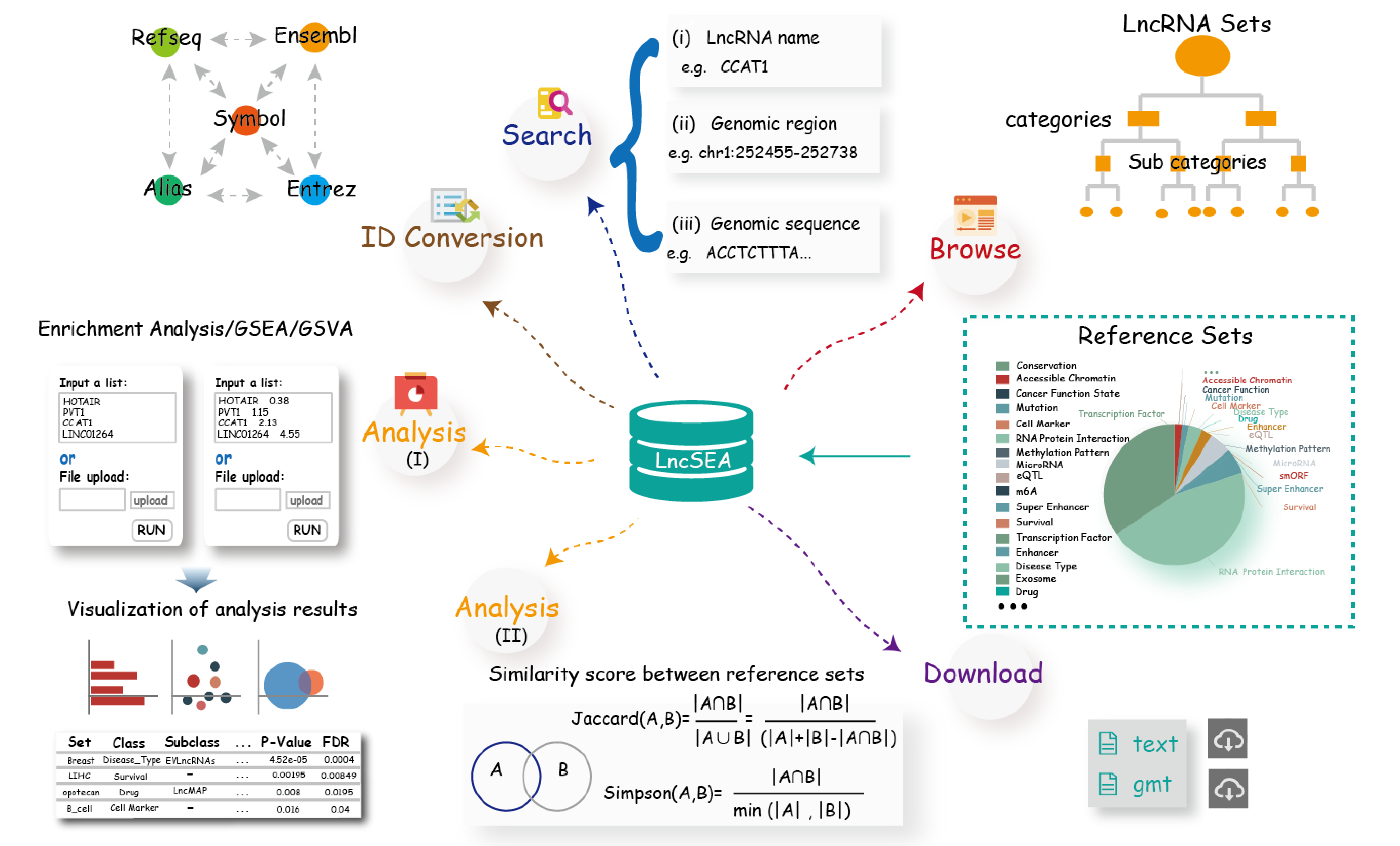
Importantly, LncSEA 2.0 provides annotation and enrichment analysis functions of lncRNA sets associated with upstream regulators and downstream targets. In addition, LncSEA 2.0 provides a user-friendly interface to search, browse and visualize detailed information about these lncRNA sets. In summary, LncSEA 2.0 is a powerful platform that provides a variety of types of lncRNA sets for users and supports lncRNA annotation and enrichment analysis functions.
LncSEA 2.0 construction process based on snakemake

Category distribution
Increases in data from LncSEA 1.0 to LncSEA 2.0
 Main functions and operational
guidelines
Main functions and operational
guidelines
 Sister Projects
Sister Projects
TF-Marker : A TF-Marker database for human
TcoFbase : Document a large number of available resources of mammals TcoFs
ENdb : an experimentally supported enhancer database for human and mouse
SEanalysis : a web tool for super-enhancer associated regulatory analysis
ATACdb : A comprehensive human chromatin accessibility database
KnockTF : a comprehensive human gene expression profile database with knockdown/knockout of transcription factors
TRCirc : a resource for transcriptional regulation information of circRNAs
TRlnc : a comprehensive database of human transcriptional regulation of lncRNAs
VARAdb : a variation annotation database for human
Publication
Chen J, Zhang J, Gao Y, Li Y, Feng C, Song C, Ning Z, Zhou X, Zhao J, Feng M, Zhang Y, Wei L, Pan Q, Jiang Y, Qian F, Han J, Yang Y, Wang Q, Li C. LncSEA: a platform for long non-coding RNA related sets and enrichment analysis. Nucleic Acids Res. 2020 Oct 12:gkaa806. doi: 10.1093/nar/gkaa806. Epub ahead of print. PMID: 33045741.