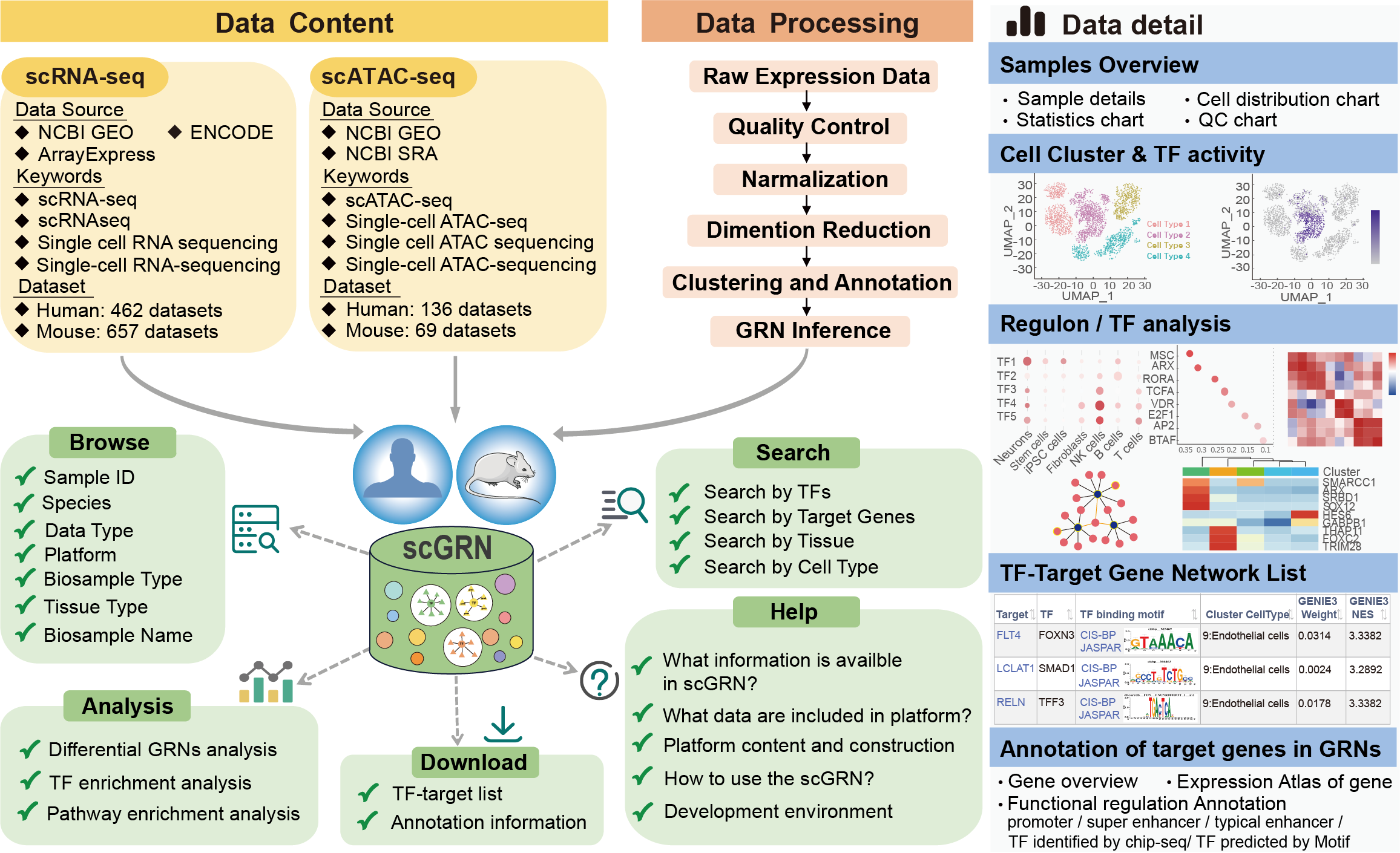
1.What information is avaiable in scGRN?
Gene regulatory networks (GRNs) formed by TFs and their downstream target genes are demonstrated to play essential roles in supervising cellular processes, such as cell division, cell growth, and cell death. Making sense of the topology and dynamics of GRNs is fundamental to interpreting the mechanisms of disease etiology and translating corresponding findings into novel therapies. Here, we have developed scGRN platform (https://bio.liclab.net/scGRN/index.php), a comprehensive single-cell multi-omics gene regulatory network platform across diverse human and mouse conditions, which aimed to record massive GRN information computationally inferred from scRNA-seq and scATAC-seq data and provide detailed epigenetics annotations of target genes.
2.What data are included in scGRN?
The current version of scGRN catalogs a total of 62,999,692 cell type-specific TF-target gene pairs inferred from 1324 single-cell samples, which covers 160 tissues/cell lines. These samples have been manually curated from NCBI GEO/SRA, ENCODE, Arrayexpress databases and include multiple single-cell sequencing platforms. To provide a better user experience, scGRN has offered multiple types of visualization methods for GRN related information and detailed functional annotation of promoters, super enhancers, typical enhancers in TF binding region. Furthermore, we have implemented three online analysis tools, that is, differential network analysis, TF enrichment analysis and pathway downstream analysis. scGRN is a user-friendly platform to query, analyze and visualize information associated with GRNs.
3.Platform content and construction
scGRN is a comprehensive resource for scGRNs with the largest samples and the most comprehensive annotation information. We provide a convenient platform for researchers to explore cell type-specific GRNs and GRNs-associated analyses. Our effort to establish this platform is of great value for researchers to explore potential functions and regulatory mechanisms in disease and biological processes.
4.How to use the scGRN?
URL:https://bio.liclab.net/scGRN/browse.php
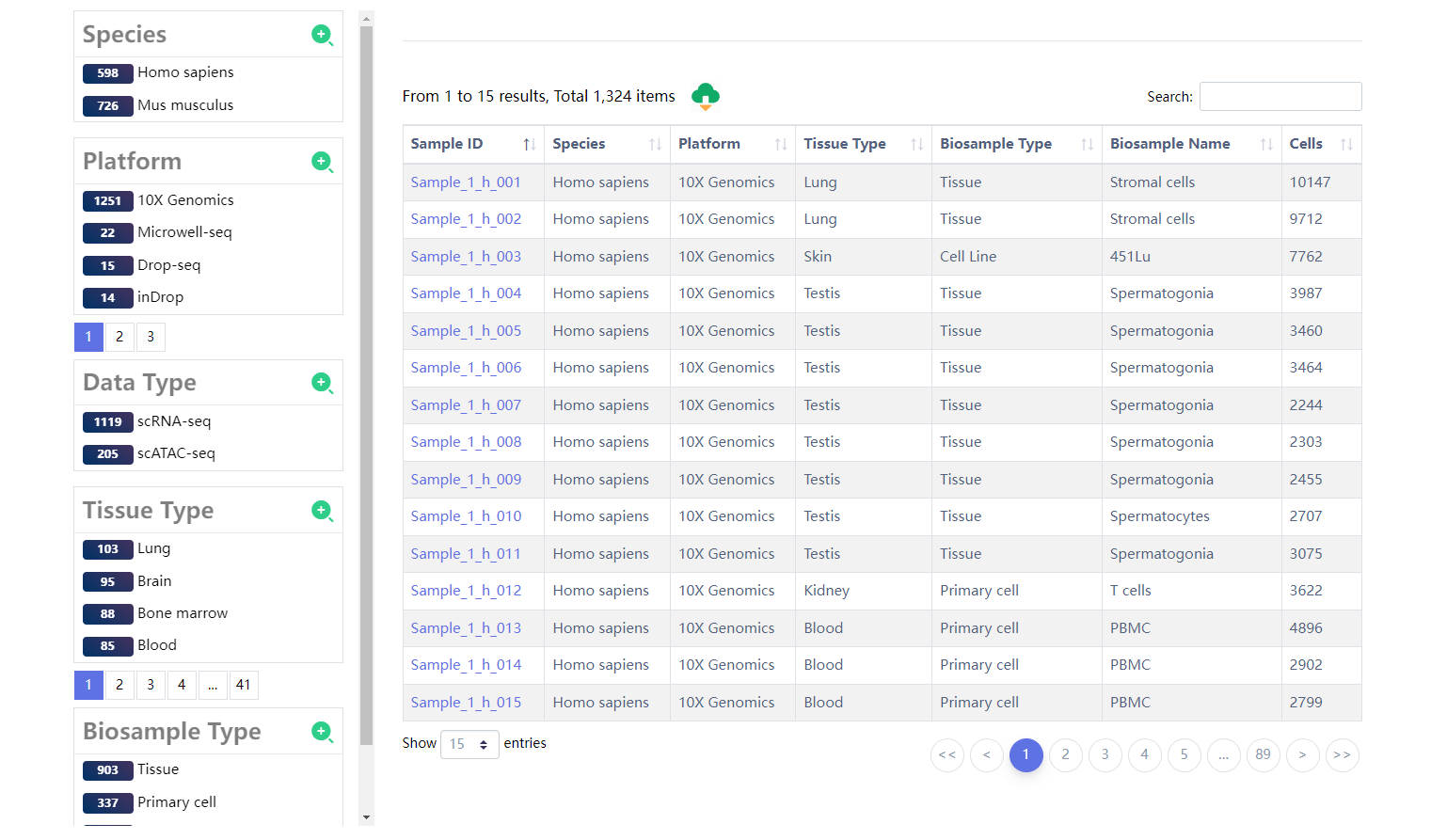
4.1 Browse
The "Browse" page is organized as an interactive and alphanumerically sortable table that allows users to quickly browse samples and customize filters through "Species", "Platform","Data Type", "Tissue Type" and "Biosample Type" in the left panel. To view the details for a given sample, users only need to click on the "Sample ID" in the right panel.
URL:https://bio.liclab.net/scGRN/search.php
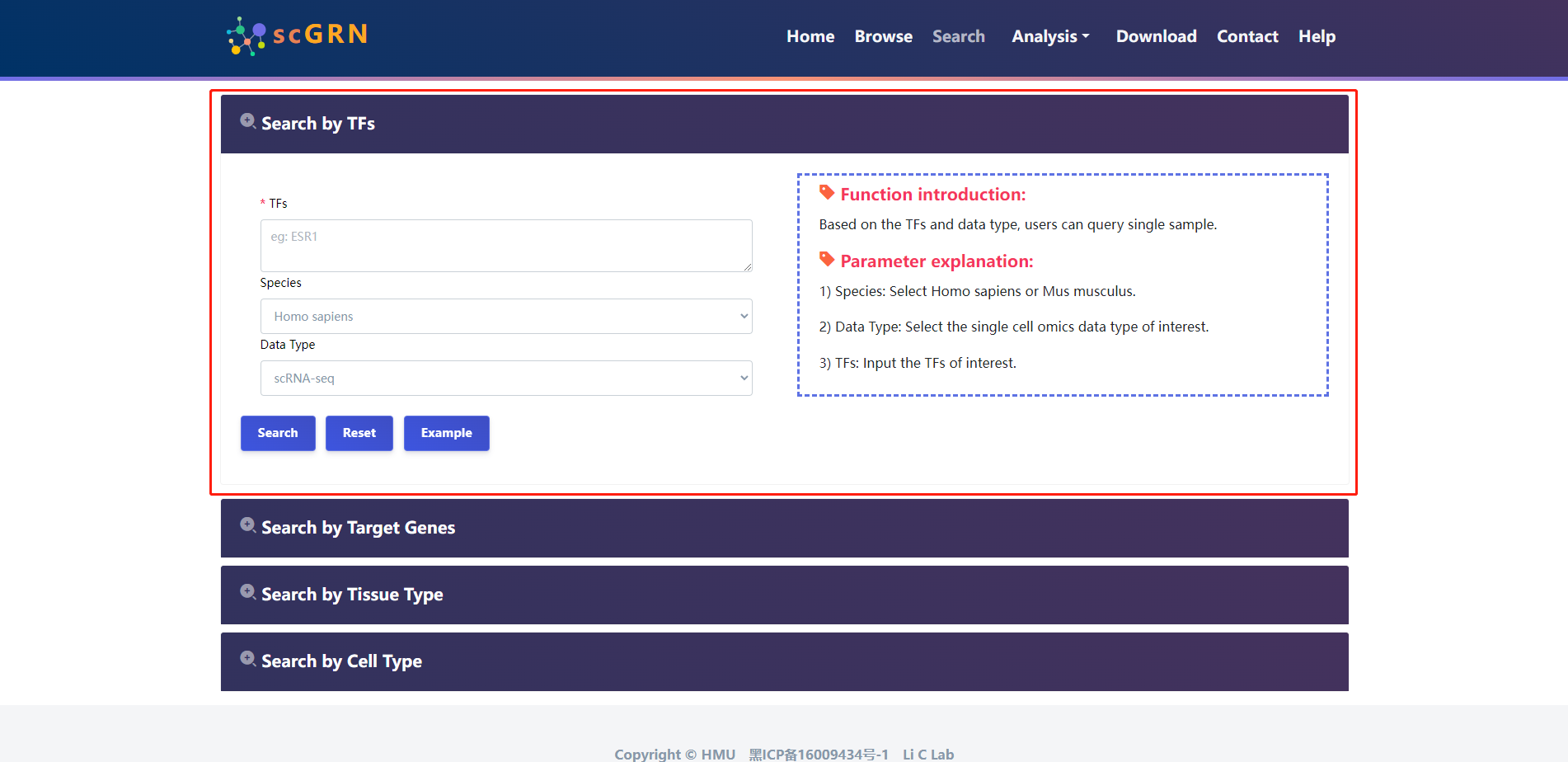
4.2 Search
scGRN provides four query methods for searching scGRNs information.
4.2.1 Search by TFs
The users select species, Data type and input TFs of interest.
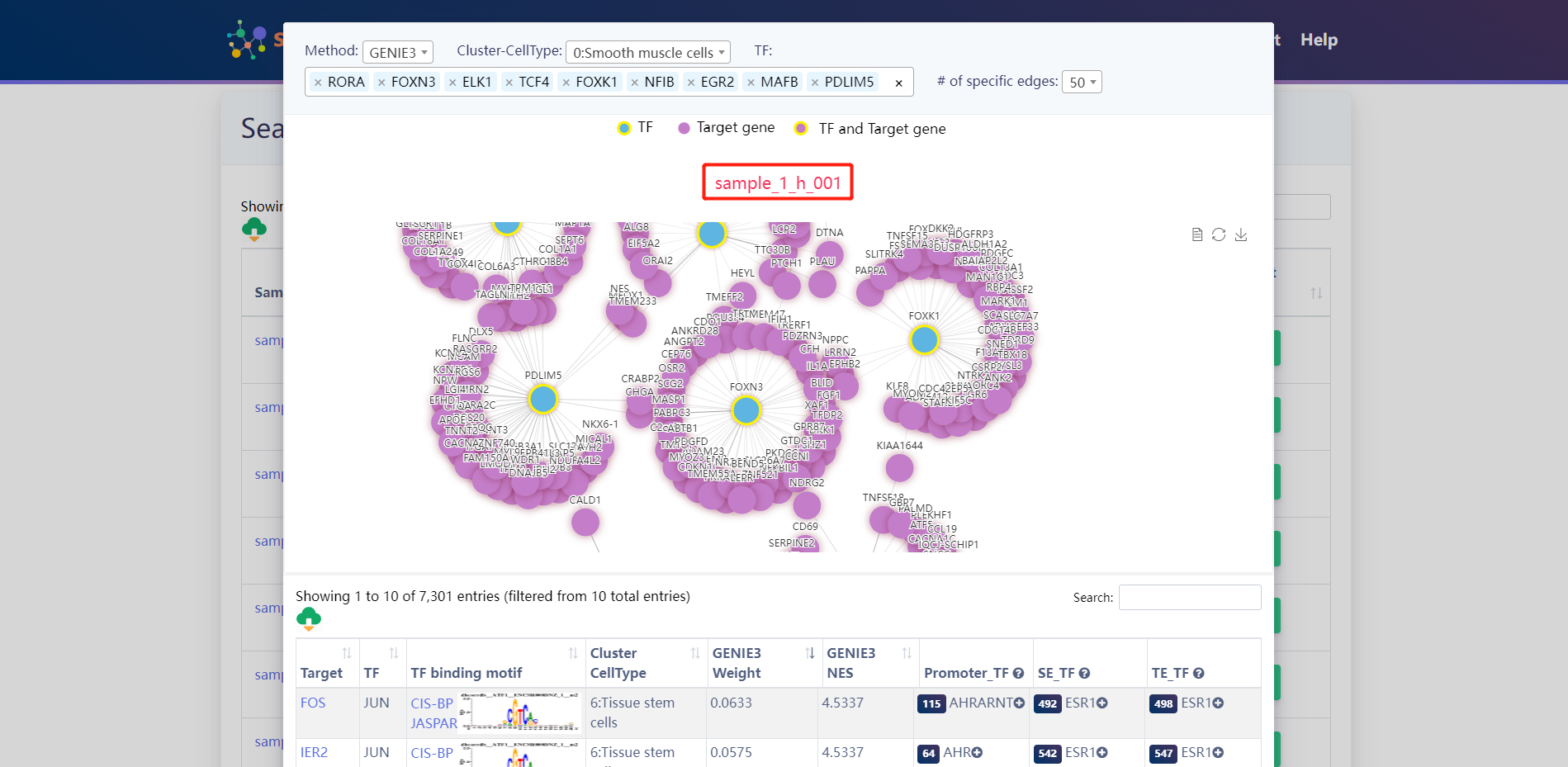
Search result of TFs for (TCF4、RORA、FOXN3、ELK1、FOXK1、NFIB、MAFB、PDLIM5、EGR2)
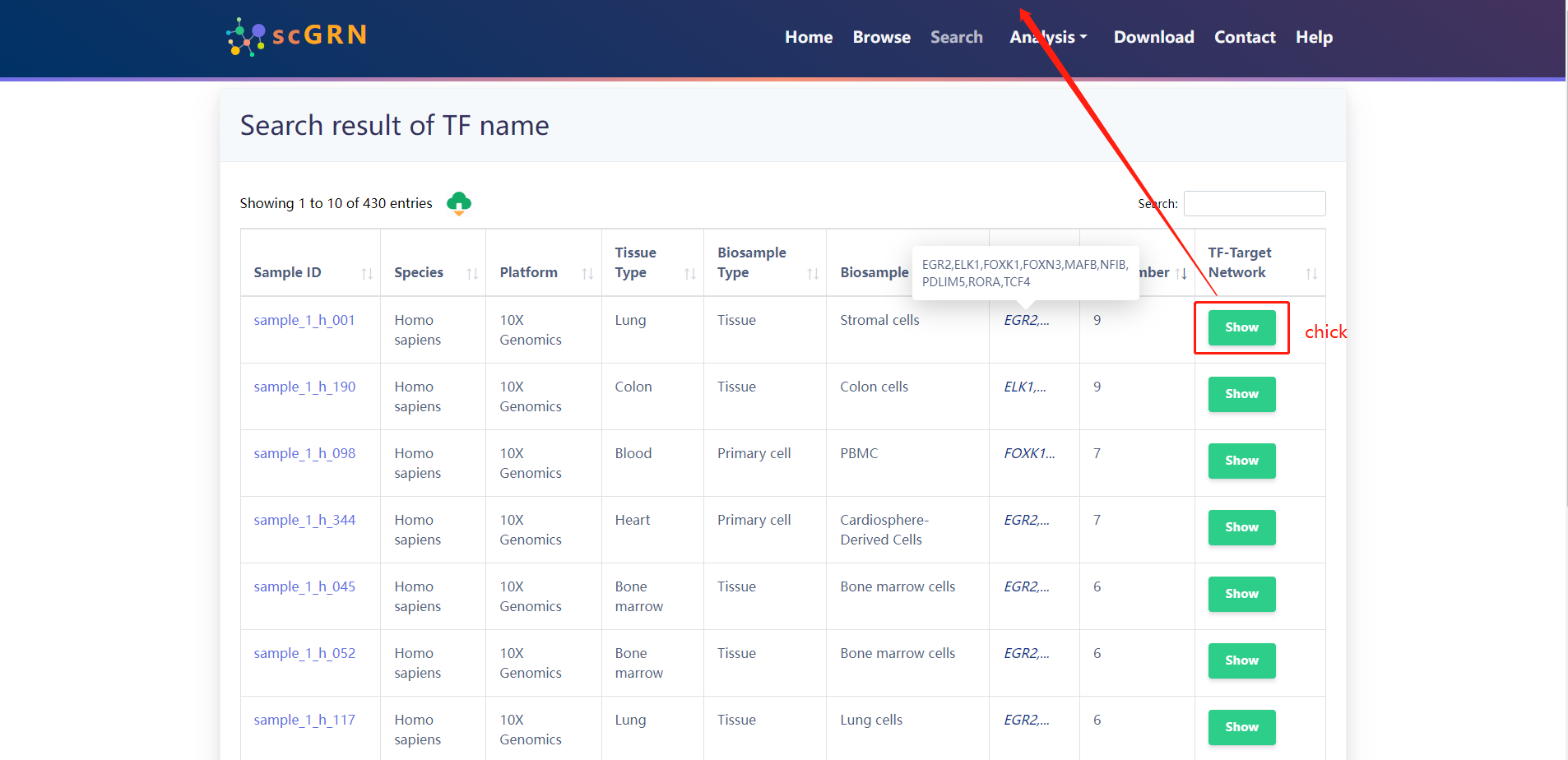
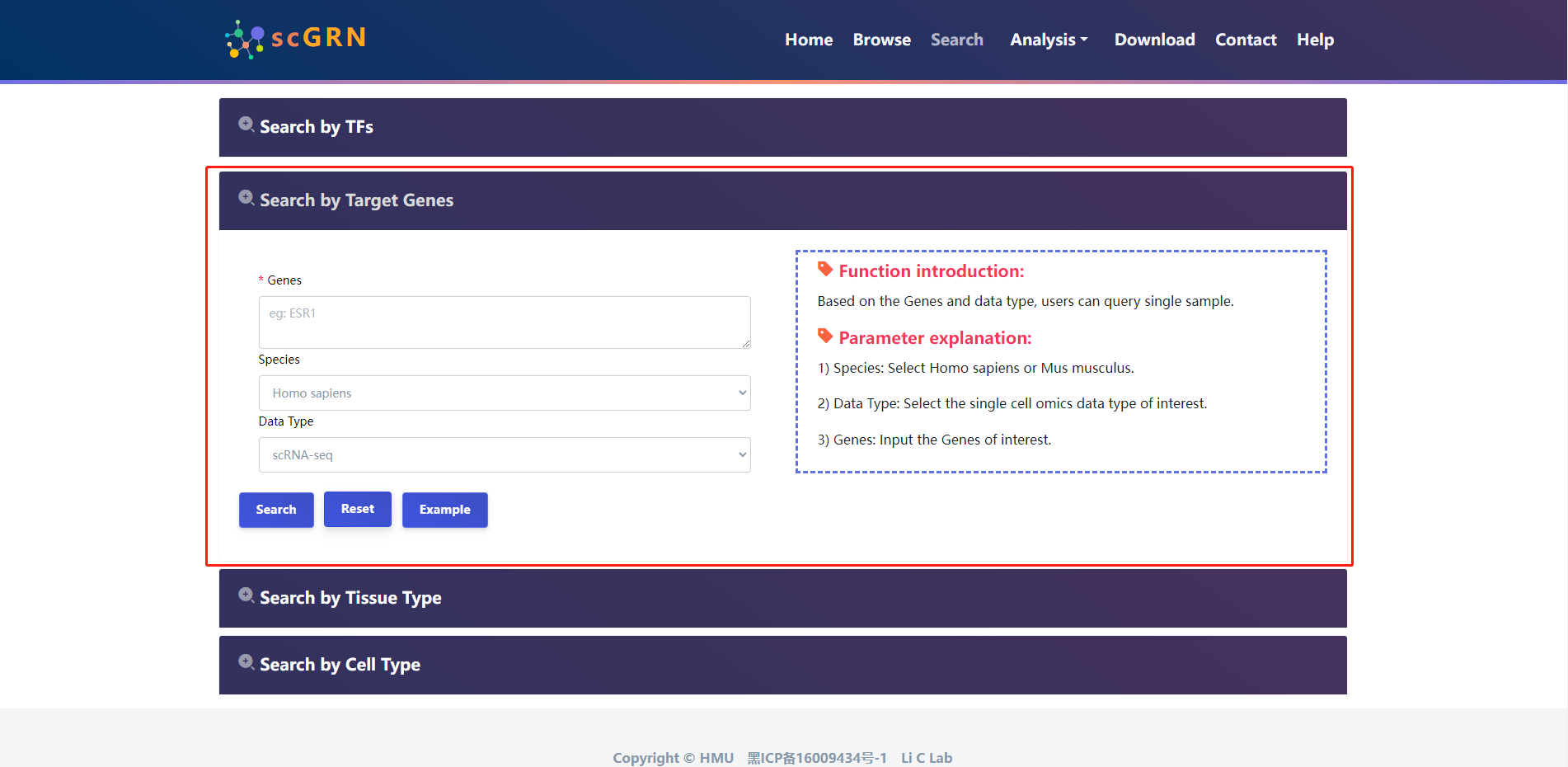
4.2.2 Search by Target Genes
The users select species and input target genes of interest.
 Search result of Target Genes for (TCF4、RORA、FOXN3、ELK1、FOXK1、NFIB、MAFB、PDLIM5、EGR2)
Search result of Target Genes for (TCF4、RORA、FOXN3、ELK1、FOXK1、NFIB、MAFB、PDLIM5、EGR2)
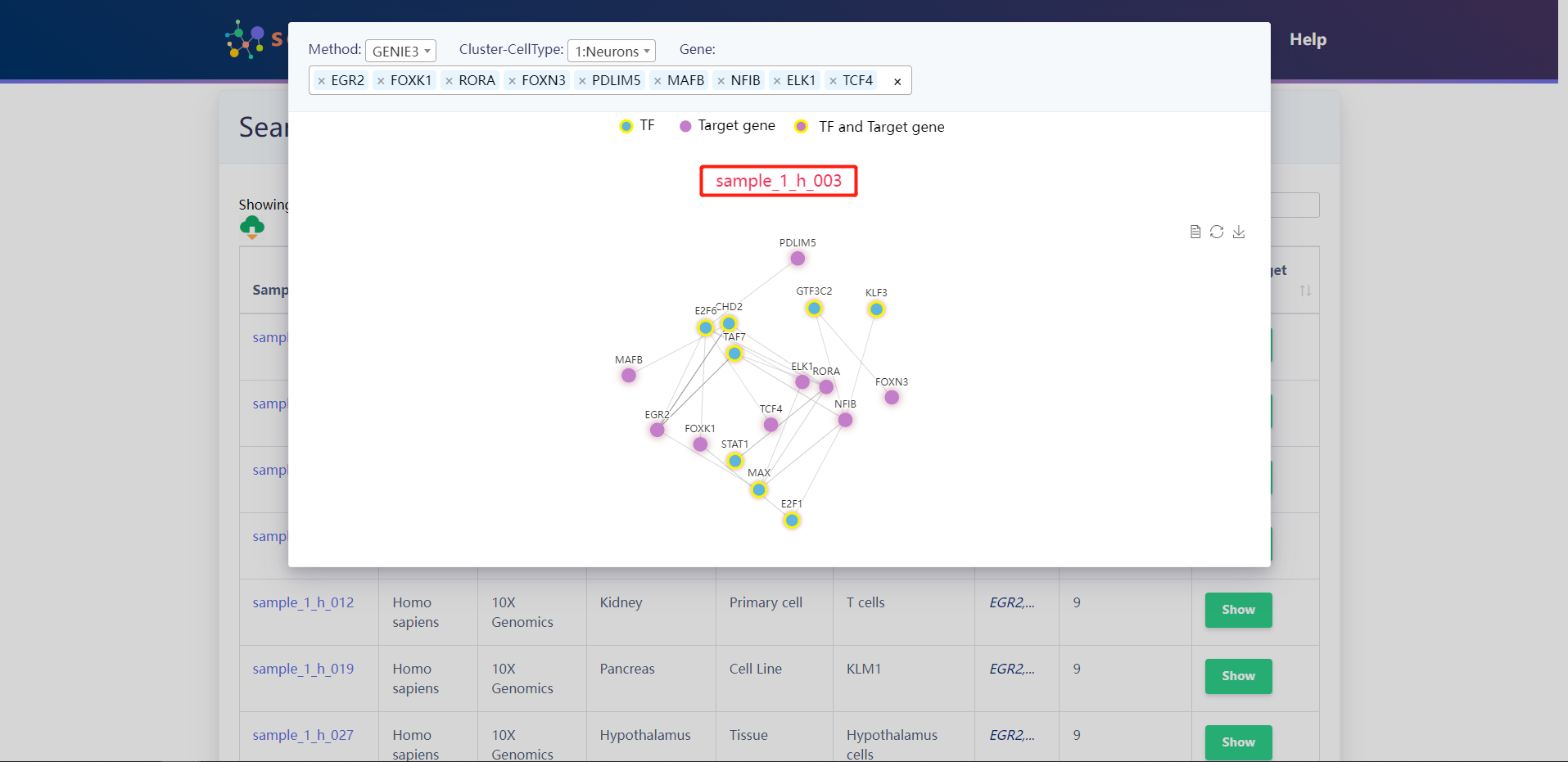
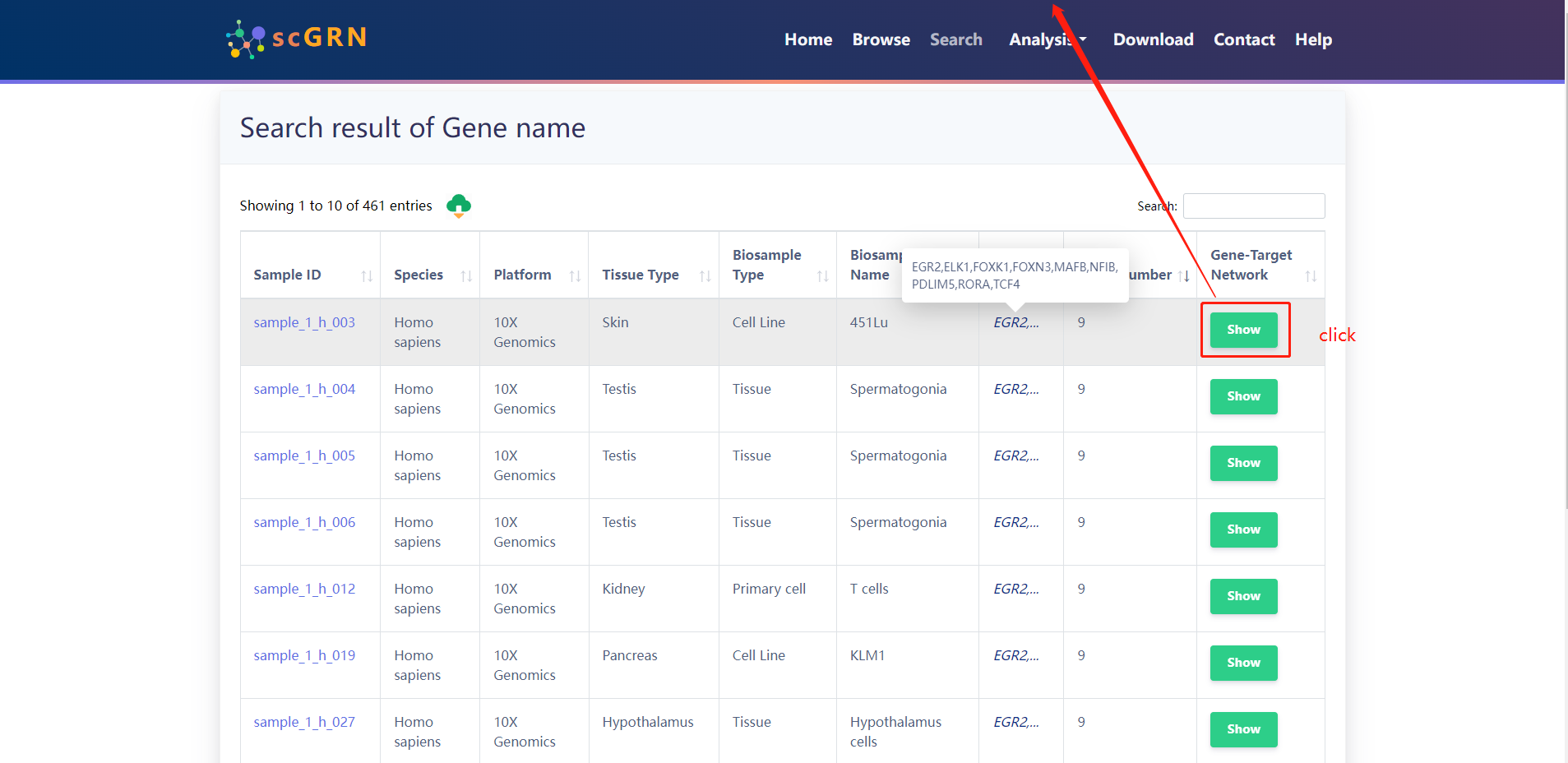
4.2.3 Search by Tissue Type
The users select species and tissue type of interest.
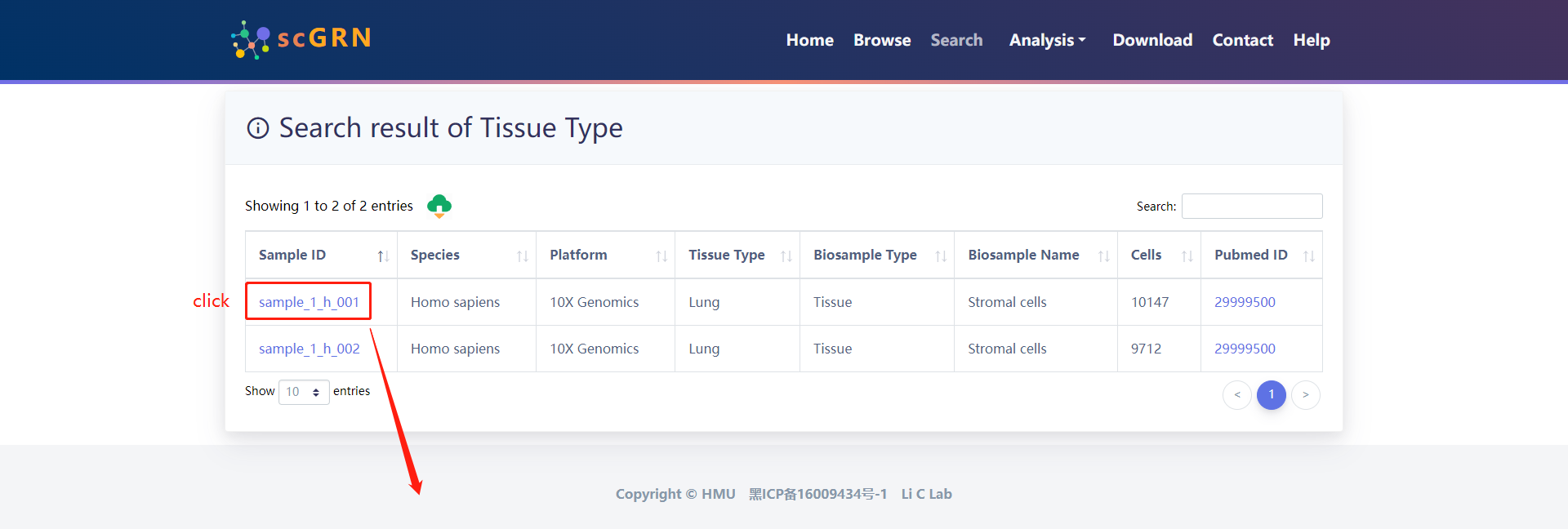
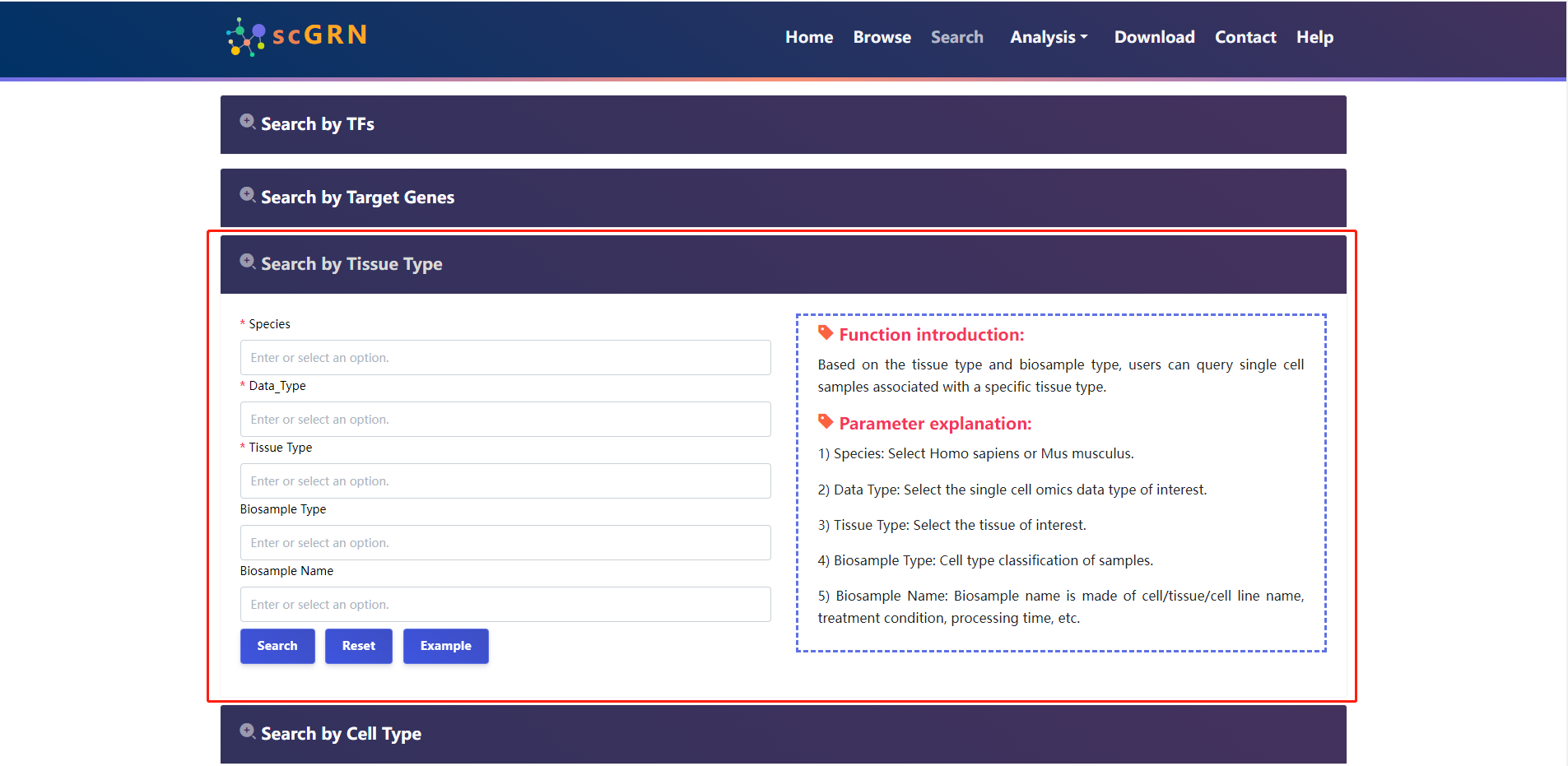 Search result of Tissue Type for example data
Search result of Tissue Type for example data
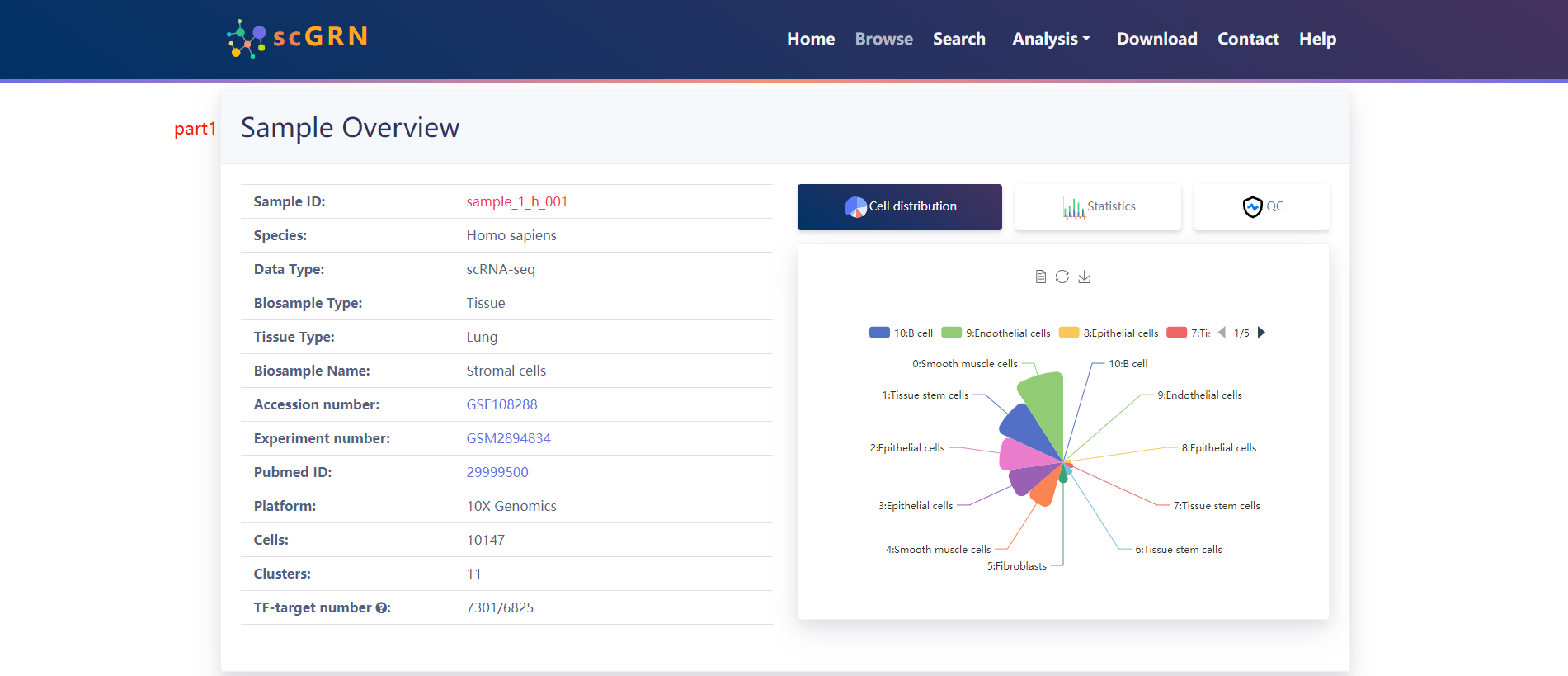
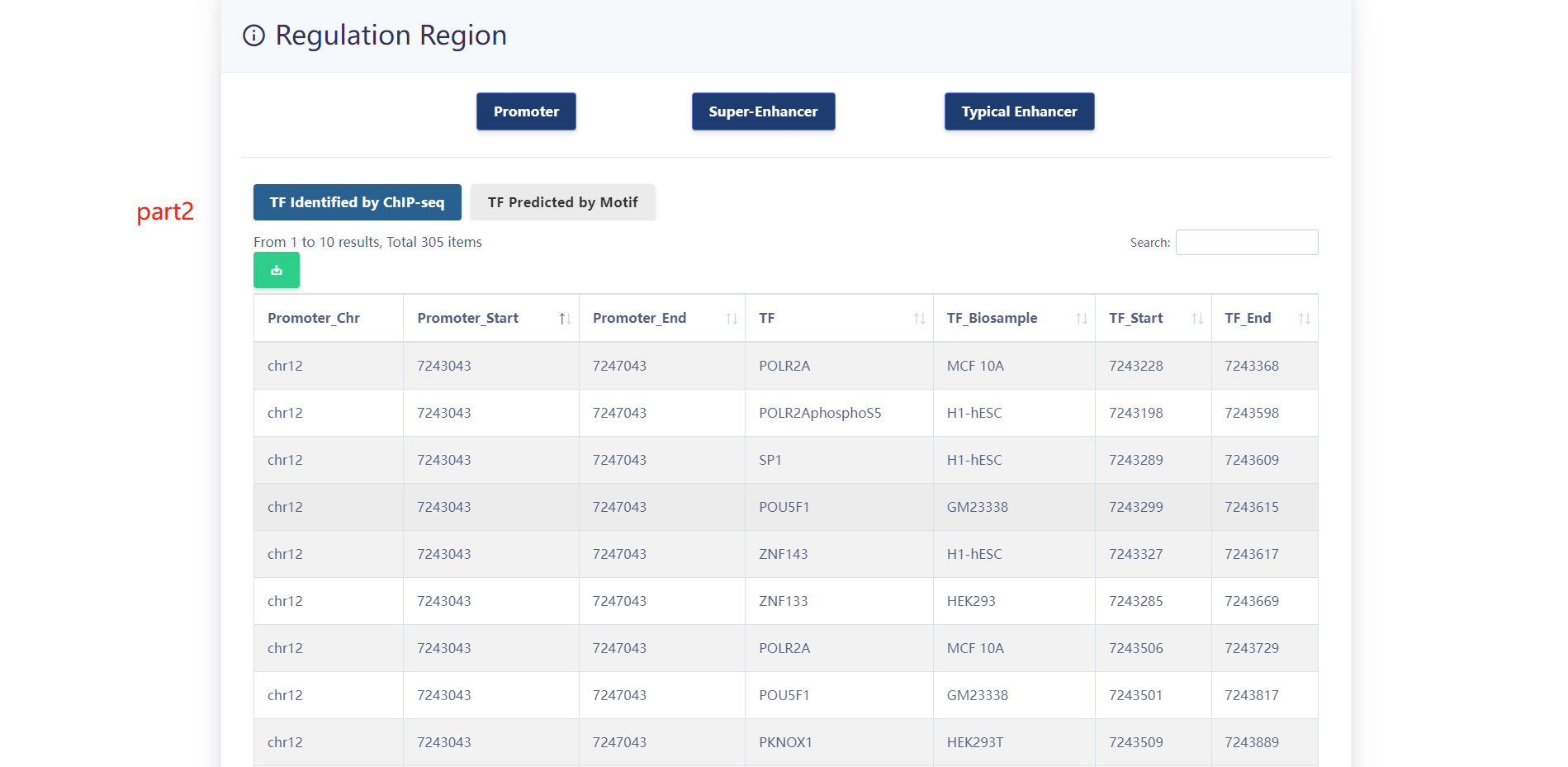
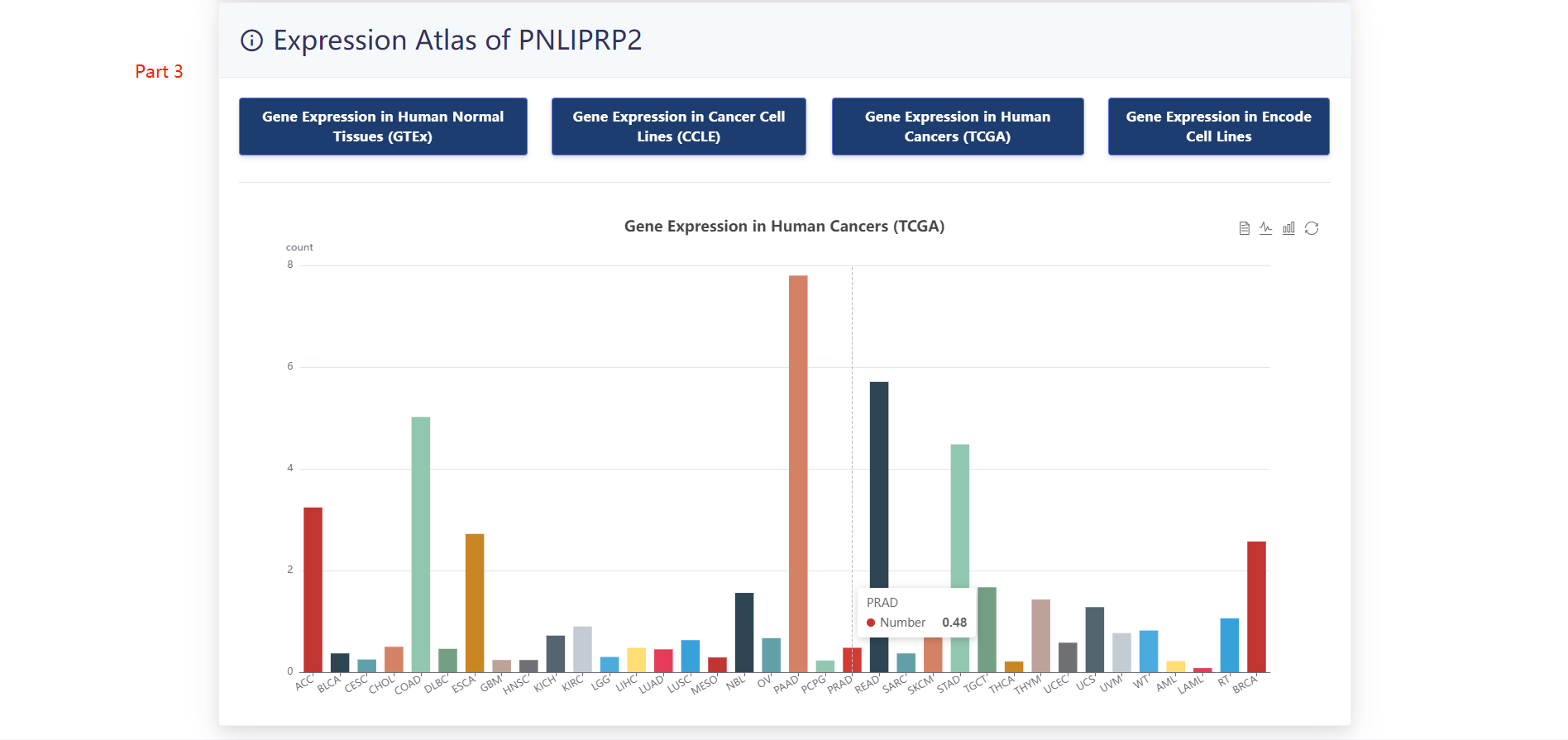
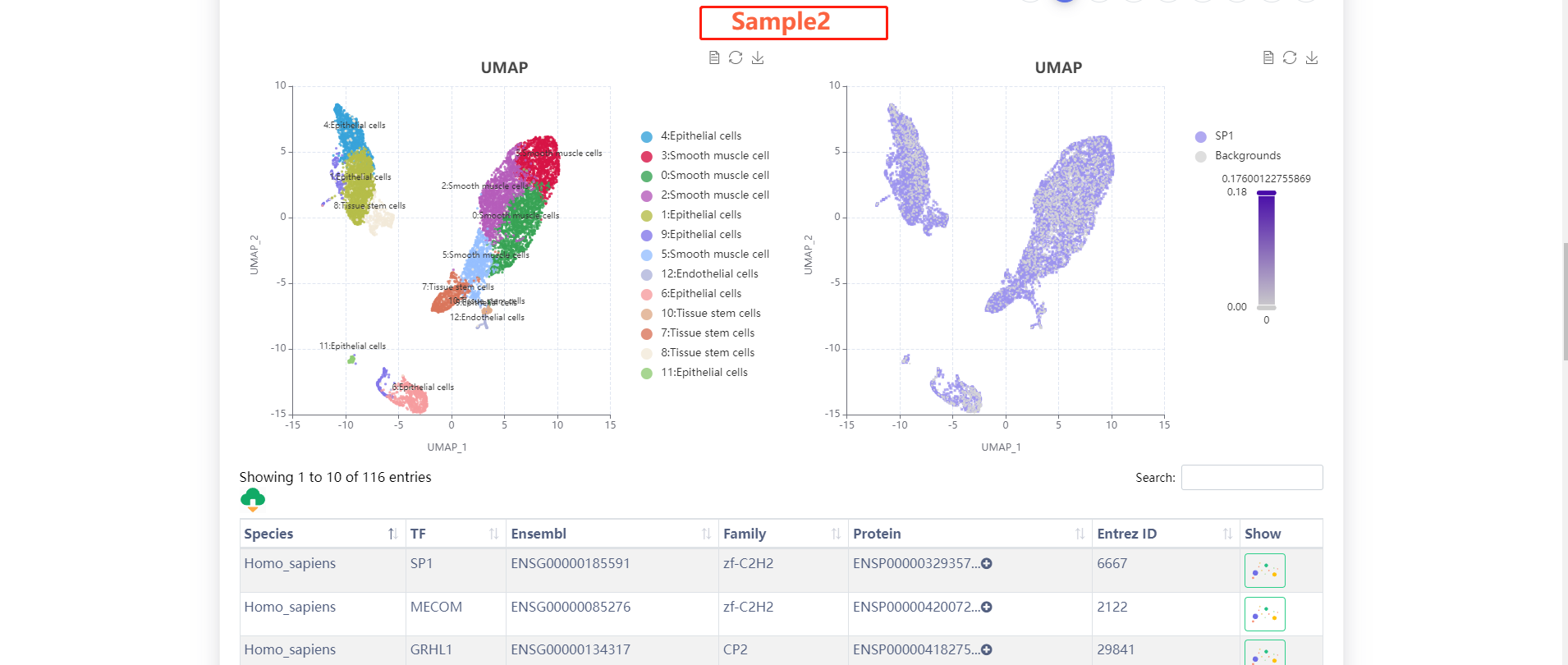
 When users choose a particular sample, three panels are presented.
When users choose a particular sample, three panels are presented.
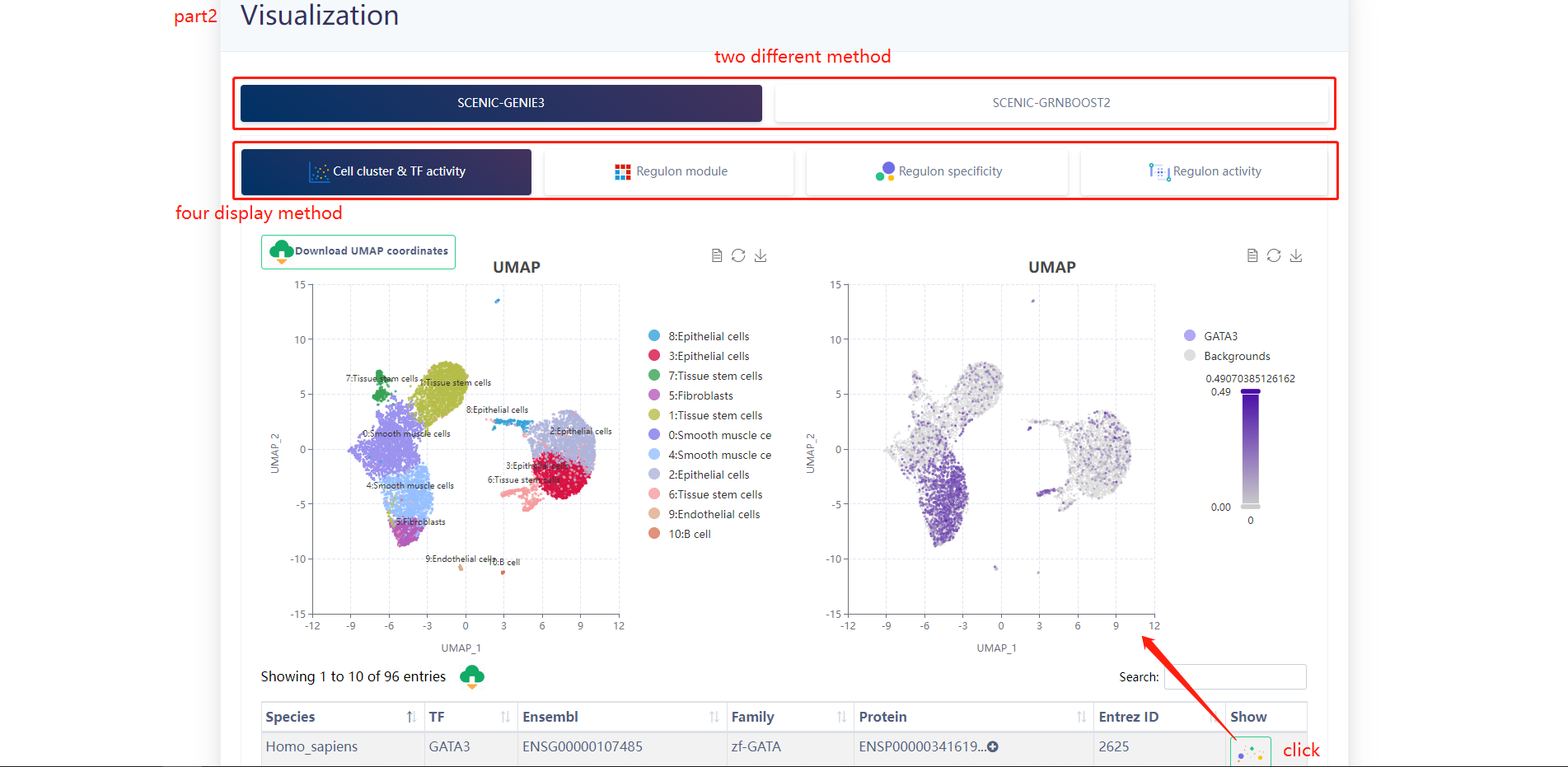

When clicking on the Target gene listed in Part3, scGRN displays the following section. Click on the PNLIPRP2 gene for a display of results.
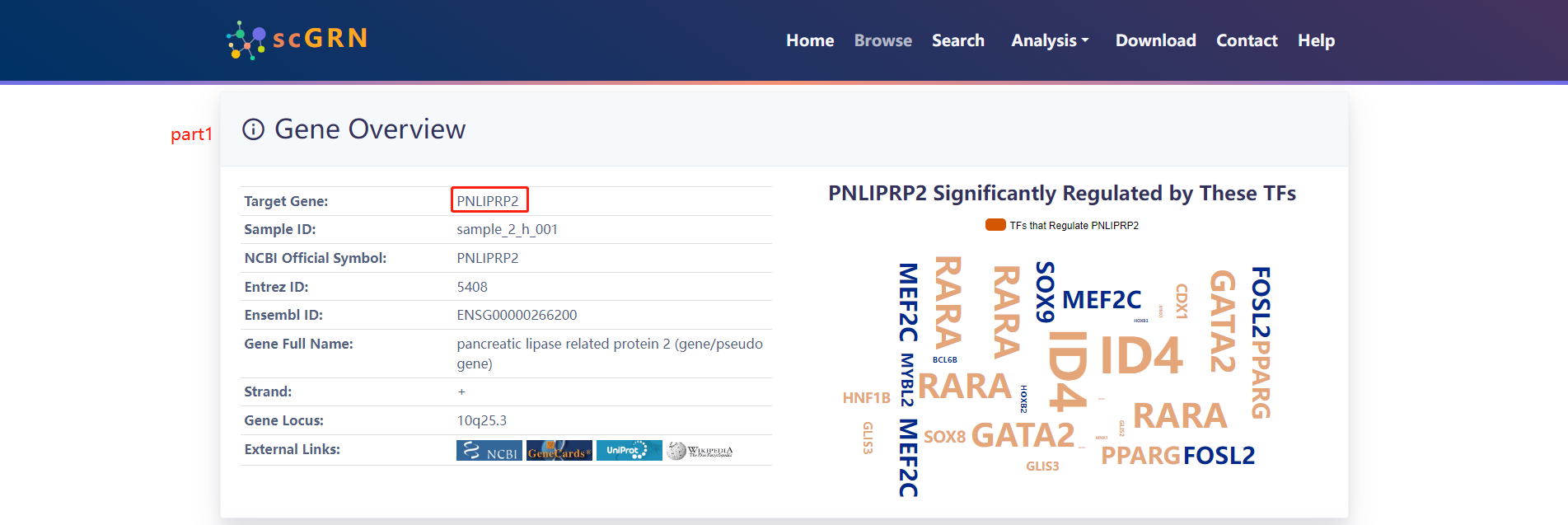
4.2.4 Search by Cell Type
The users select species and cell type of interest
 Search result of Cell Type for example data
Search result of Cell Type for example data
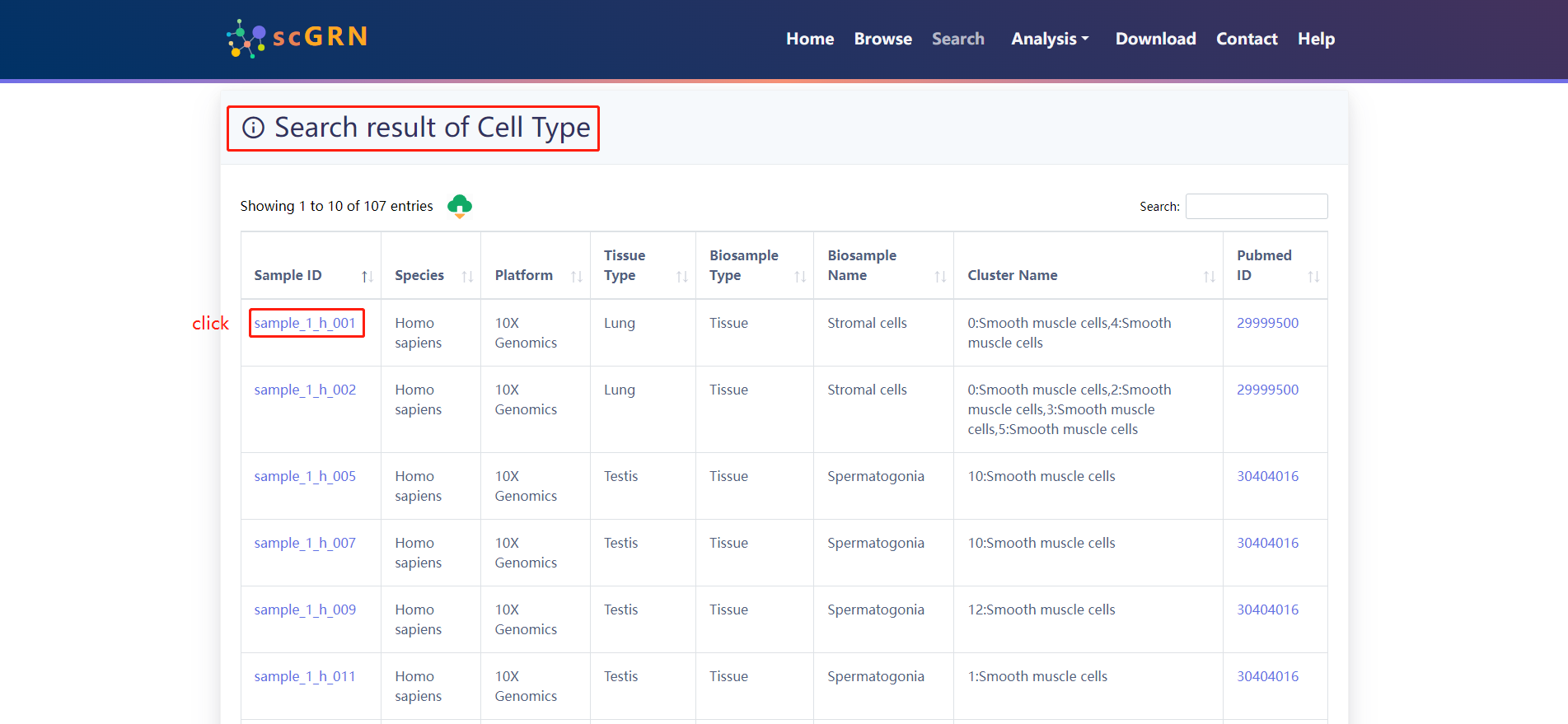 When the user selects a particular sample and click on the Sample ID, three panels similar to the type of tissue searched are displayed.
When the user selects a particular sample and click on the Sample ID, three panels similar to the type of tissue searched are displayed.
4.3 Analysis
URL:https://bio.liclab.net/scGRN/analysis/differential_network.php
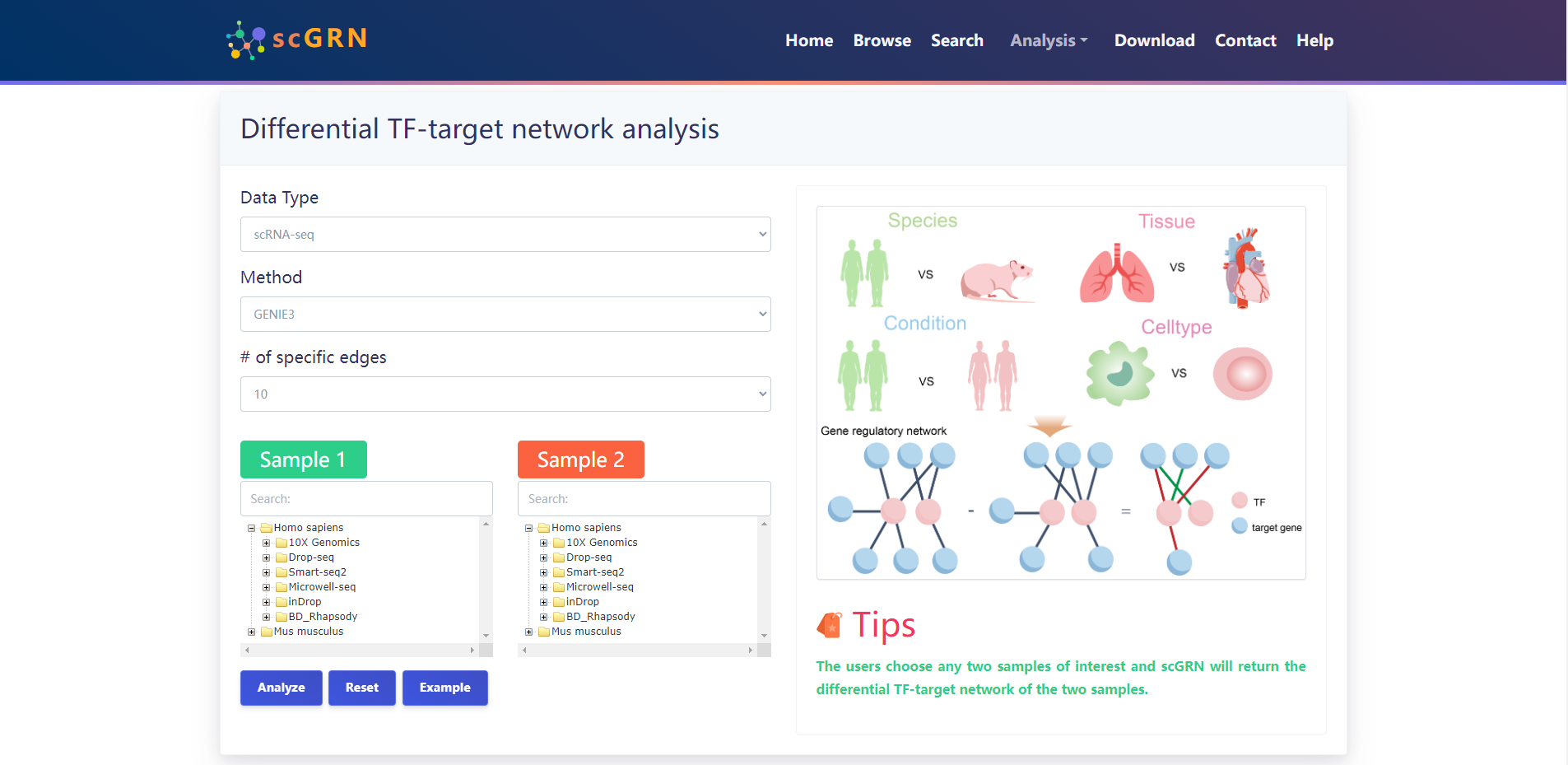
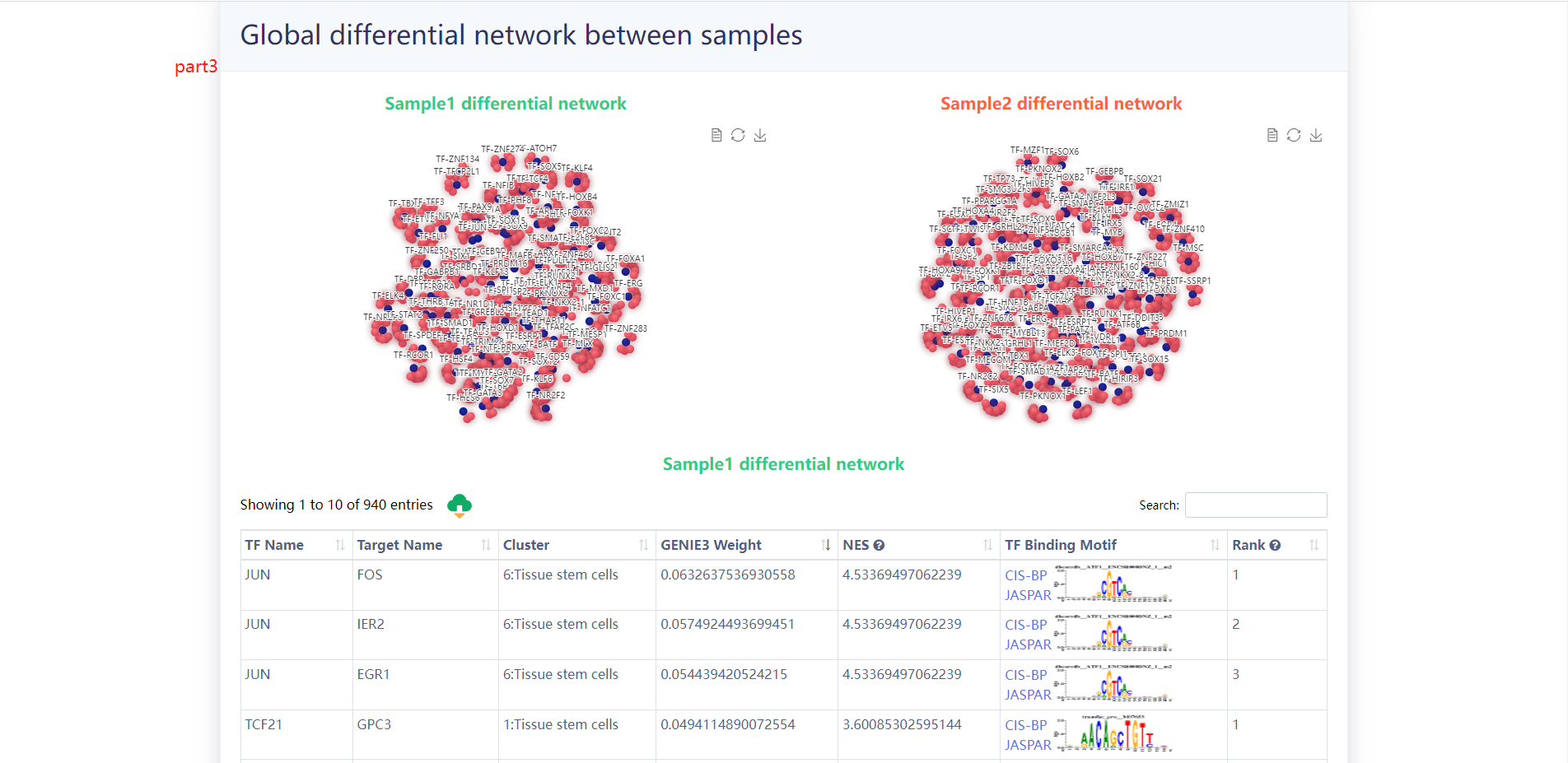
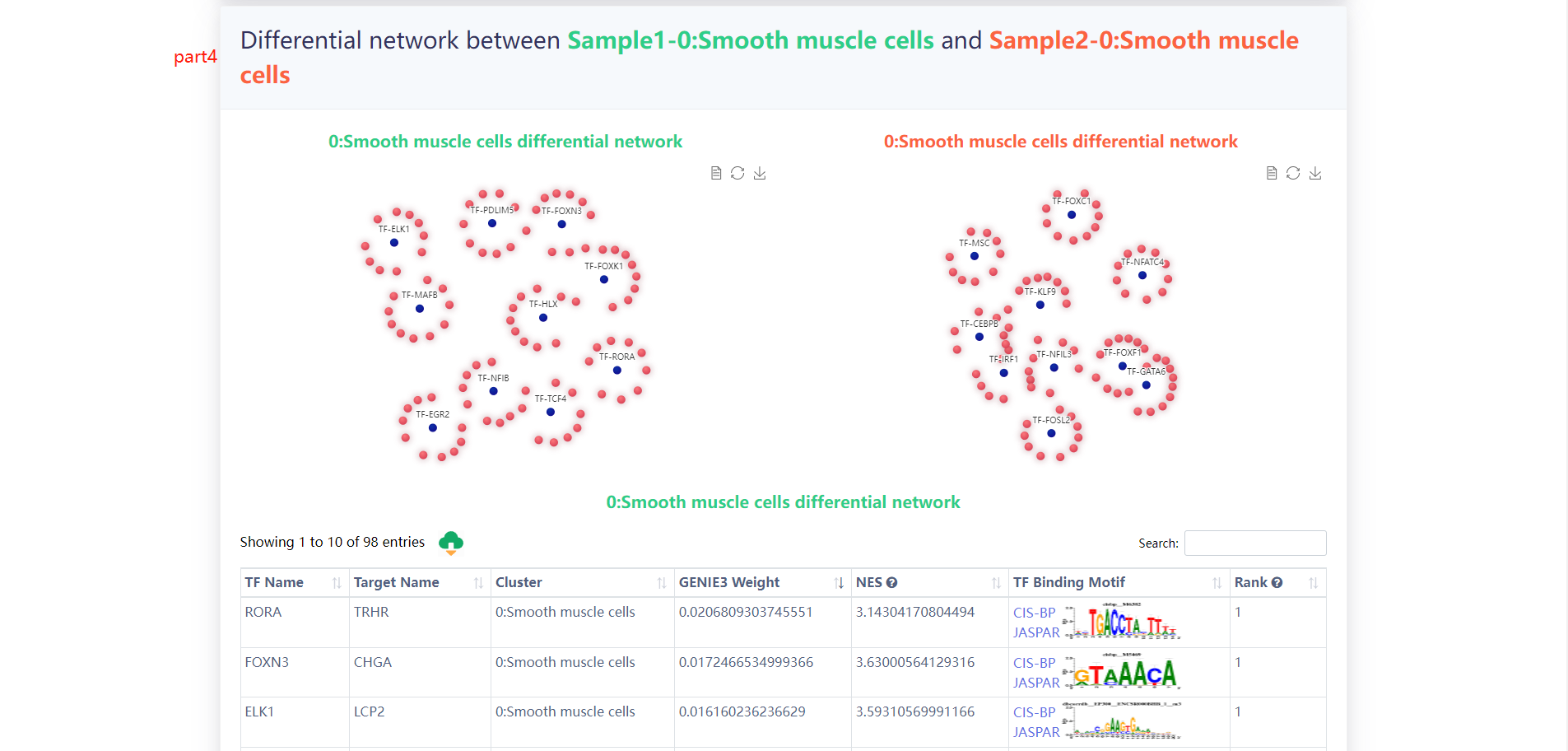
4.3.1 Differential TF-target network analysis
The users choose any two samples of interest and scGRN will return the differential TF-target network of the two samples.
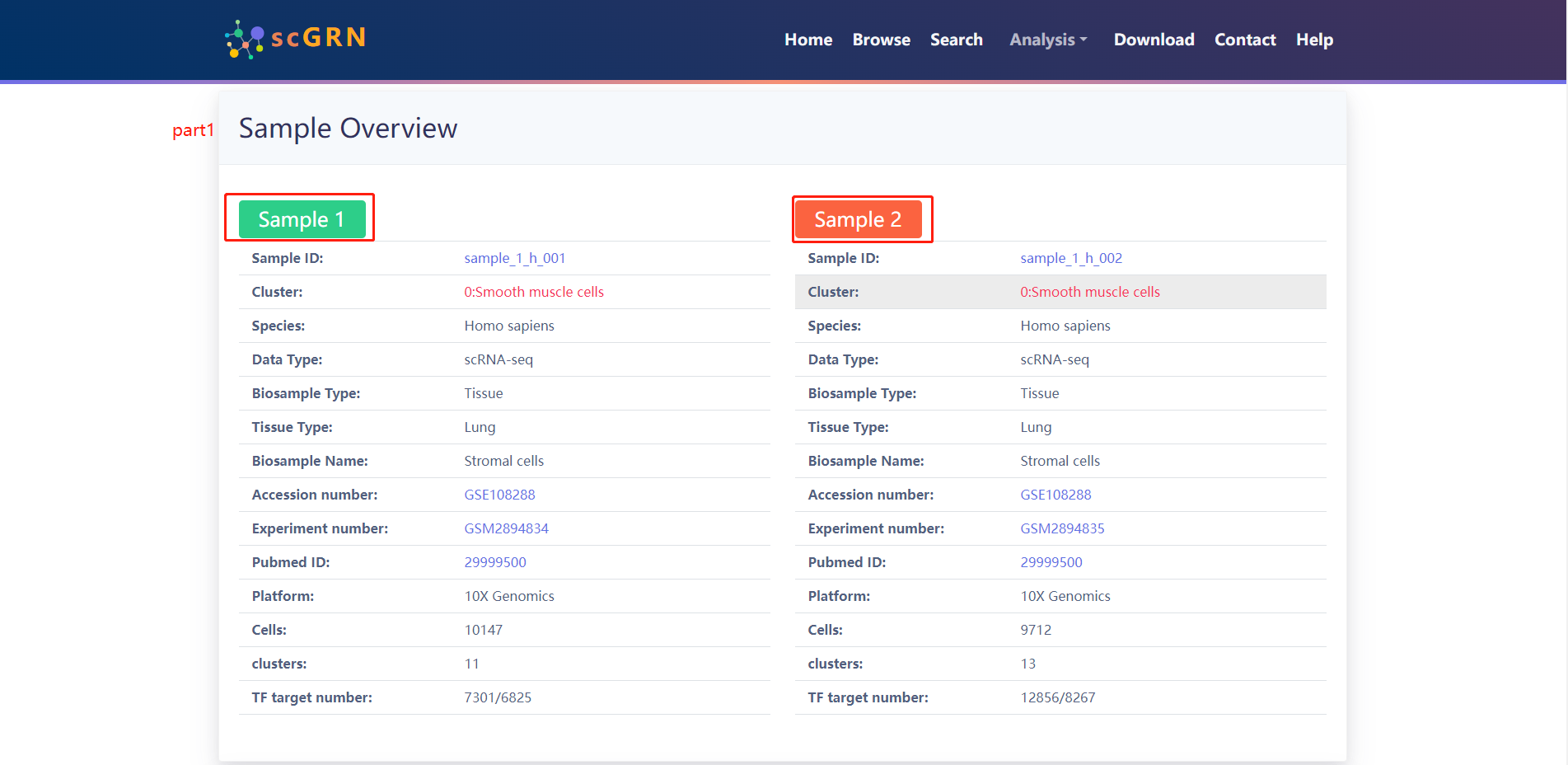
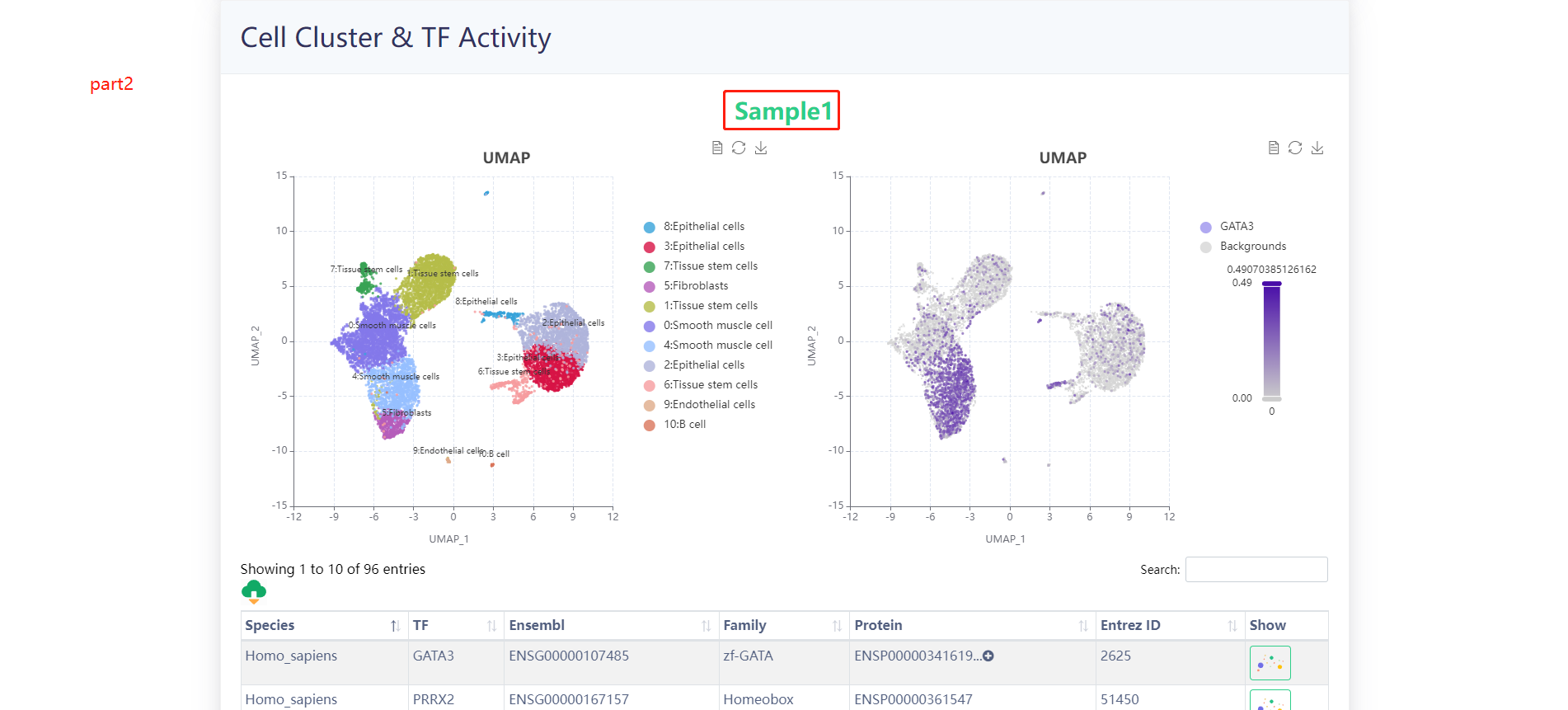
The results of the analysis of the example data are as follows.
URL:https://bio.liclab.net/scGRN/analysis/gene_set_enrichment.php
4.3.2 TF enrichment analysis
The users input a set of genes of interest, set the different thresholds needed for analysis, as well as select samples to be enriched, and scGRNbase will return the analysis results within a short time and provide the analysis results(files and graphs) for download.
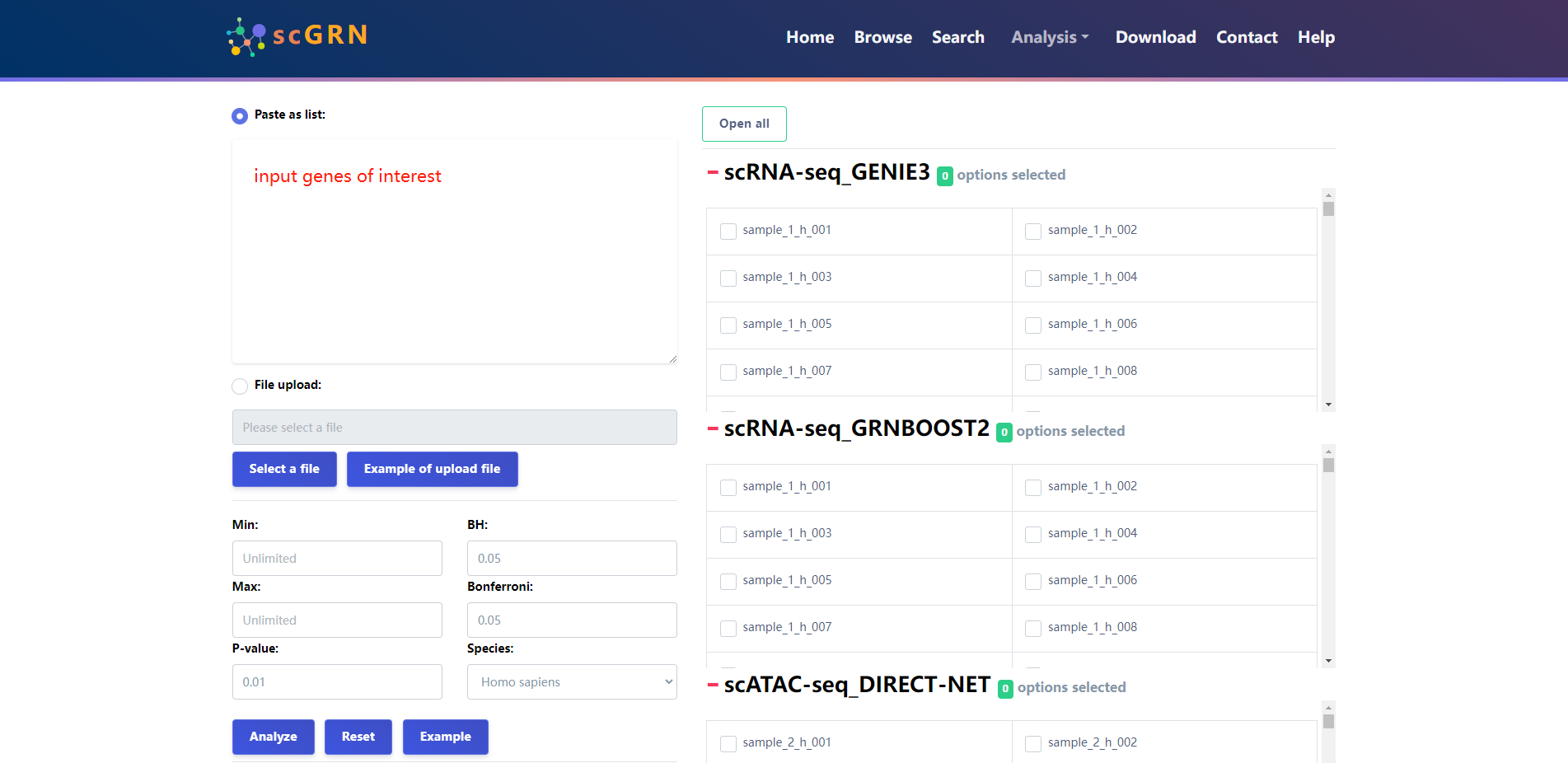
The results of the analysis of the example data are as follows.
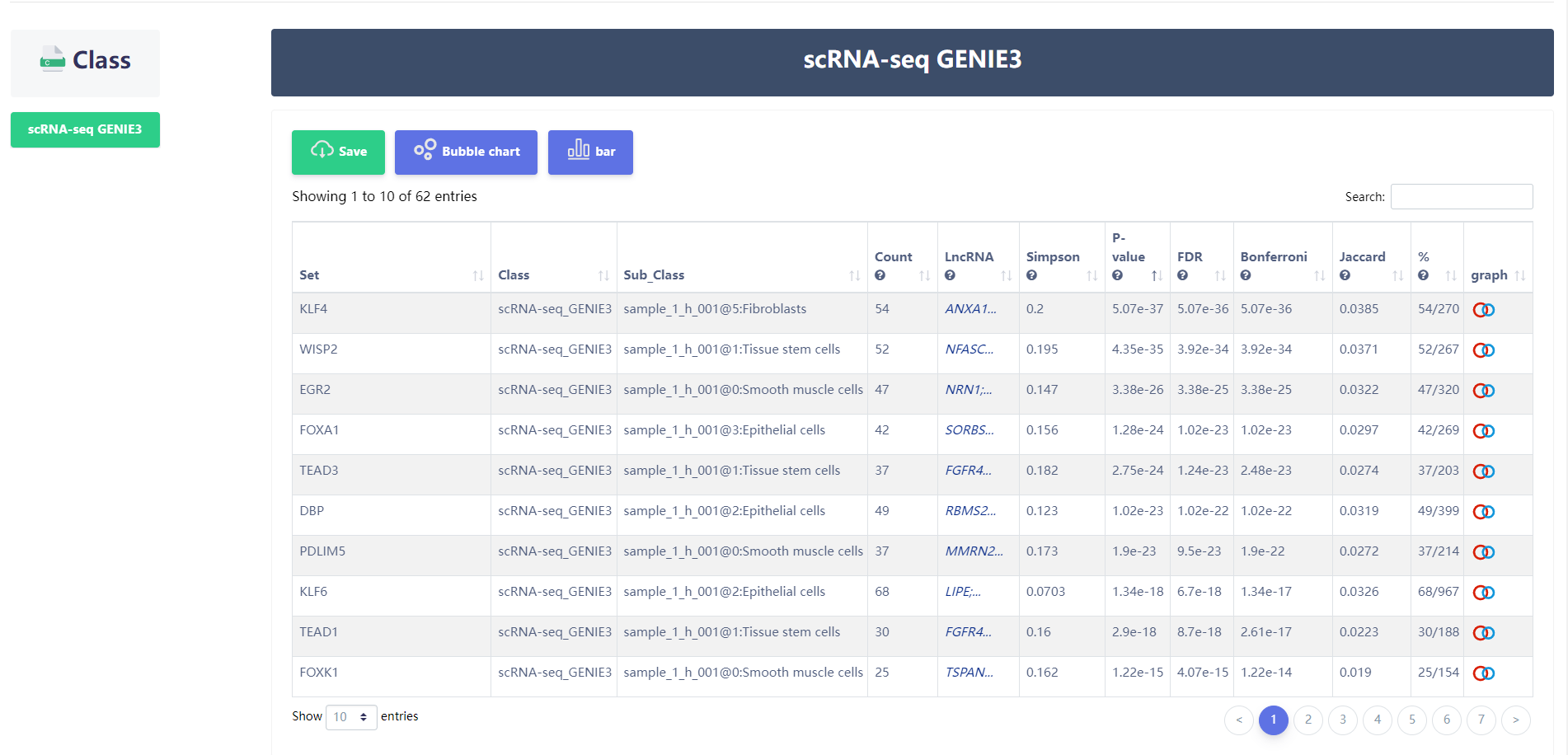
URL:https://bio.liclab.net/scGRN/analysis/pathway_enrichment_analysis.php
4.3.3 Pathway downstream analysis
The users input a set of genes of interest, and scGRNbase will identify enriched pathways using up to 10 pathway databases. In each pathway, scGRNbase will locate the terminal TF and extract the related GRNs.
The results of the analysis of the example data are as follows. Users can select the enriched pathways of interest.
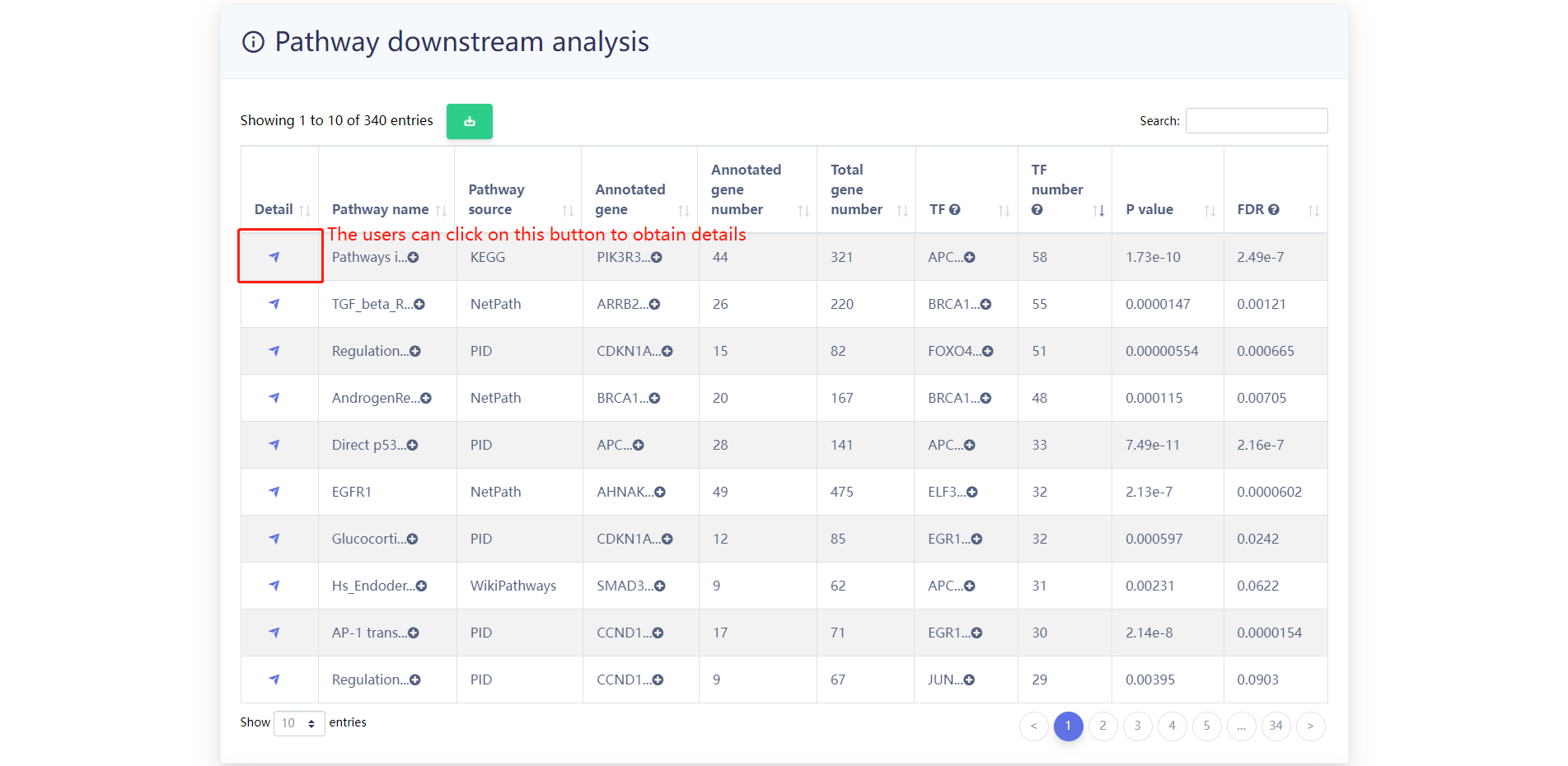
Pathway overview.
 Pathway terminal TFs related GRNs.
Pathway terminal TFs related GRNs.
URL:https://bio.liclab.net/scGRN/download.php
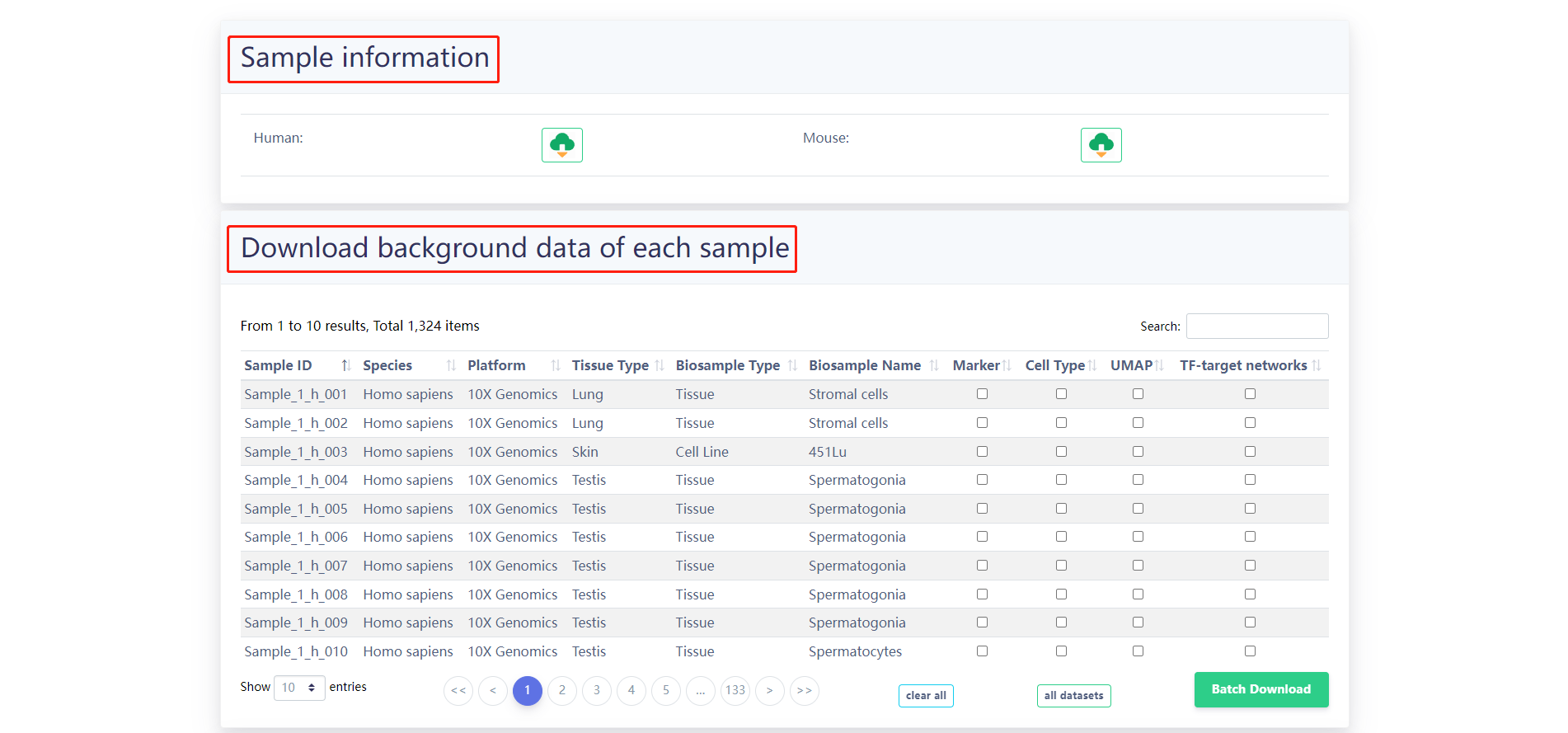
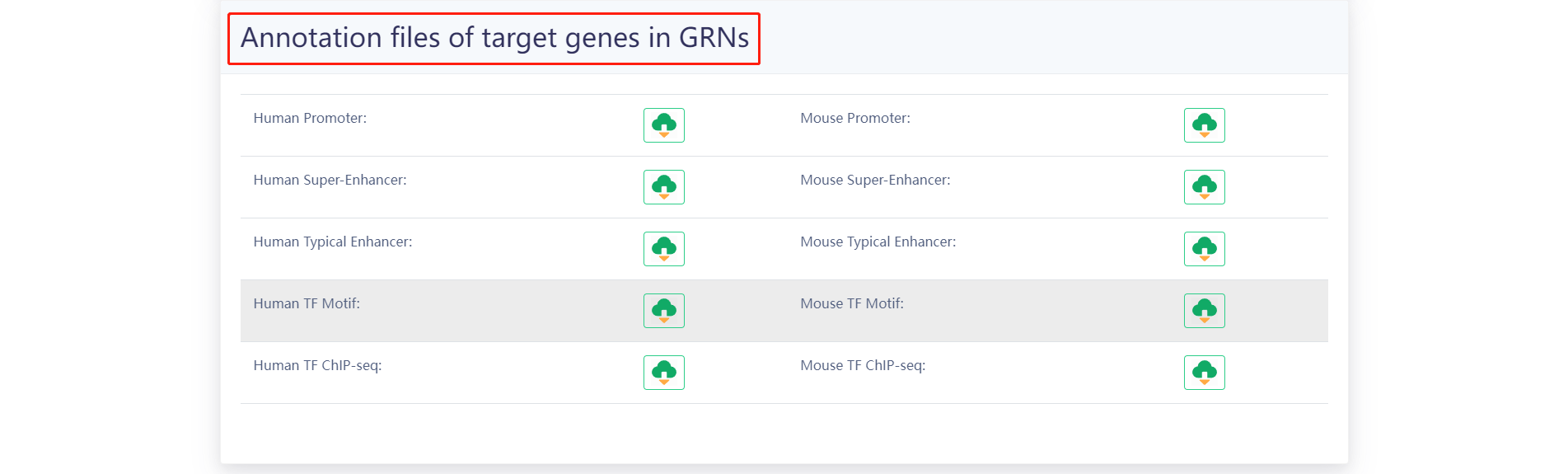
4.4 Download
The results of the GRNs for each sample as well as the annotation files are provided for download in the "Download" page. Users can quickly download associated information.
5.Development environment
The current version of scGRN was developed using MySQL 5.7.17 (http://www.mysql.com) and runs on a Linux-based Apache Web server (http://www.apache.org). We used PHP 7.0 (http://www.php.net) for server-side scripting. We designed and built the interactive interface using Bootstrap v3.3.7 (https://v3.bootcss.com) and JQuery v2.1.1 (http://jquery. com). We used ECharts (http://echarts.baidu.com) and D3 (https://d3js.org) as a graphical visualization framework, and JBrowse (http://jbrowse.org) is the genome browser framework. We recommend using a modern web browser that supports the HTML5 standard such as Firefox, Google Chrome, Safari, Opera or IE 9.0+ for the best display.
The scGRN platform is freely available to the research community using the web link (https://bio.liclab.net/scGRN/index.php). Users are not required to register or login to access features in the platform.
6.Material disclaimer
The materials and frameworks used by scGRN are shared by the network and do not contain intellectual property infringement. If there is any infringement, please write to us and we will change it in time.