CRCdb
The comprehensive human core transcriptional regulatory circuit database
What is CRCdb?
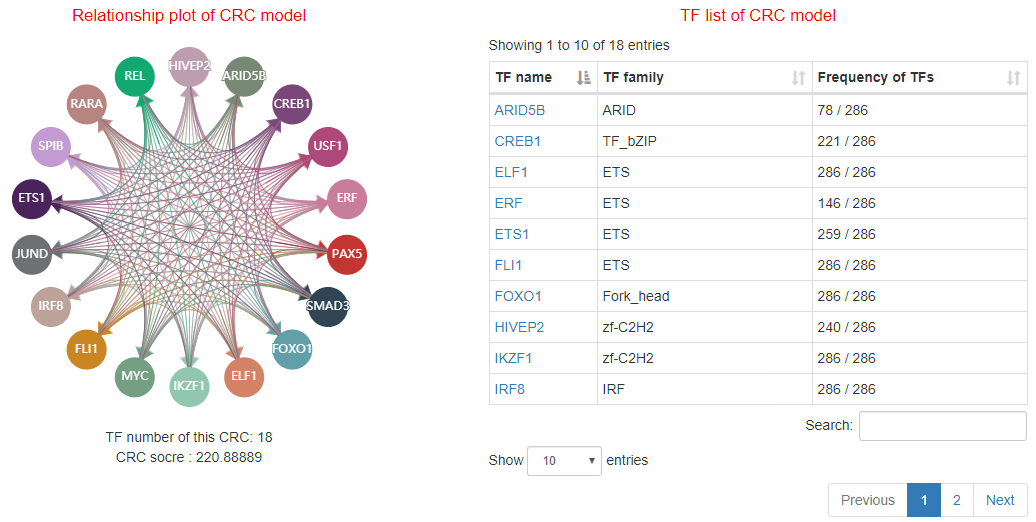
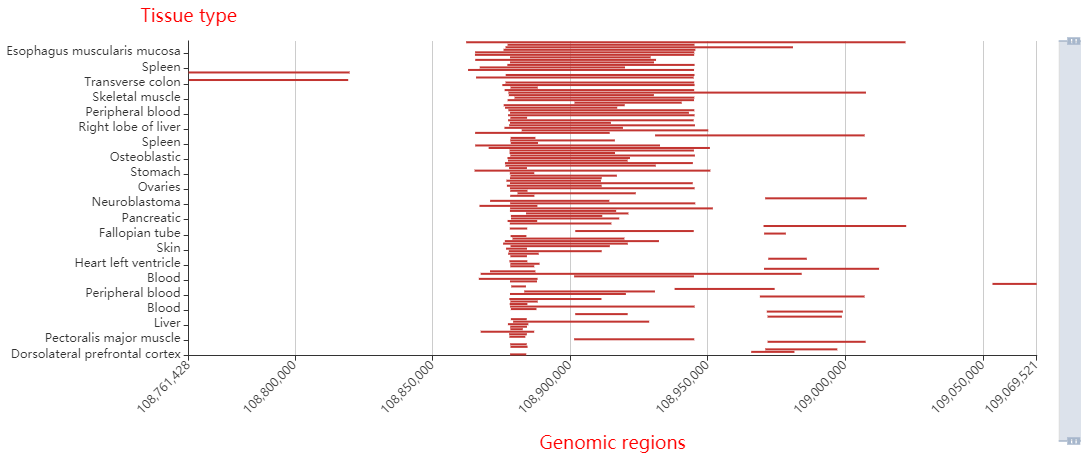
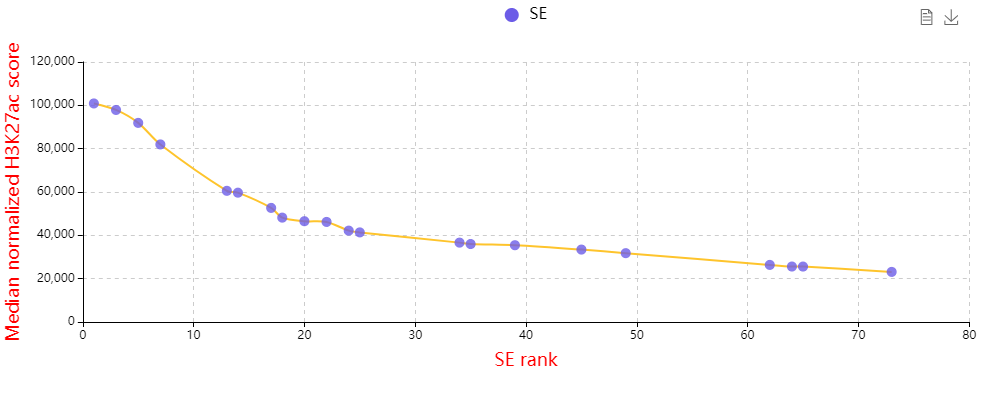
Core transcriptional regulatory circuit (CRC) is comprised of a group of interconnected auto-regulating transcription factors (TFs) forming loops, and core TFs in CRCs have been proved highly valuable for cell-type-specific transcriptional regulation in healthy and disease cells. They can be computationally inferred on the base of enhancer clusters called super-enhancers (SEs).
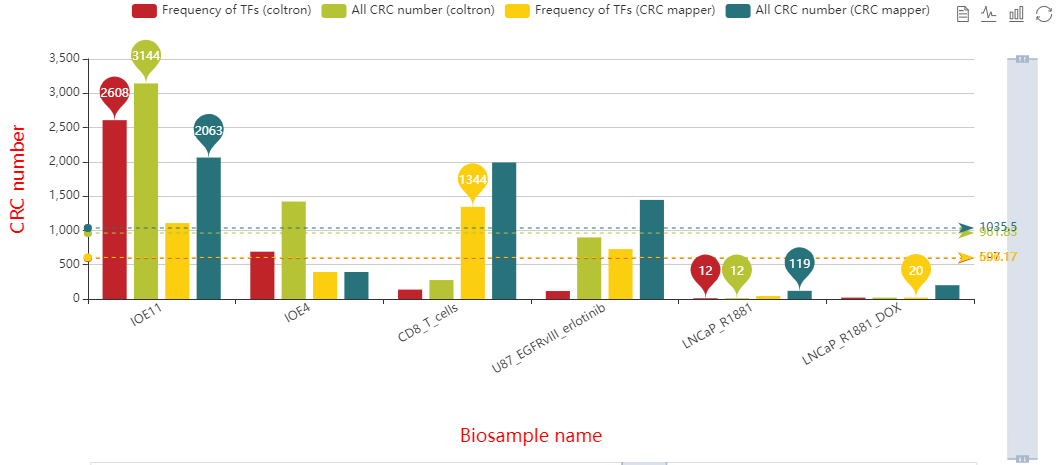
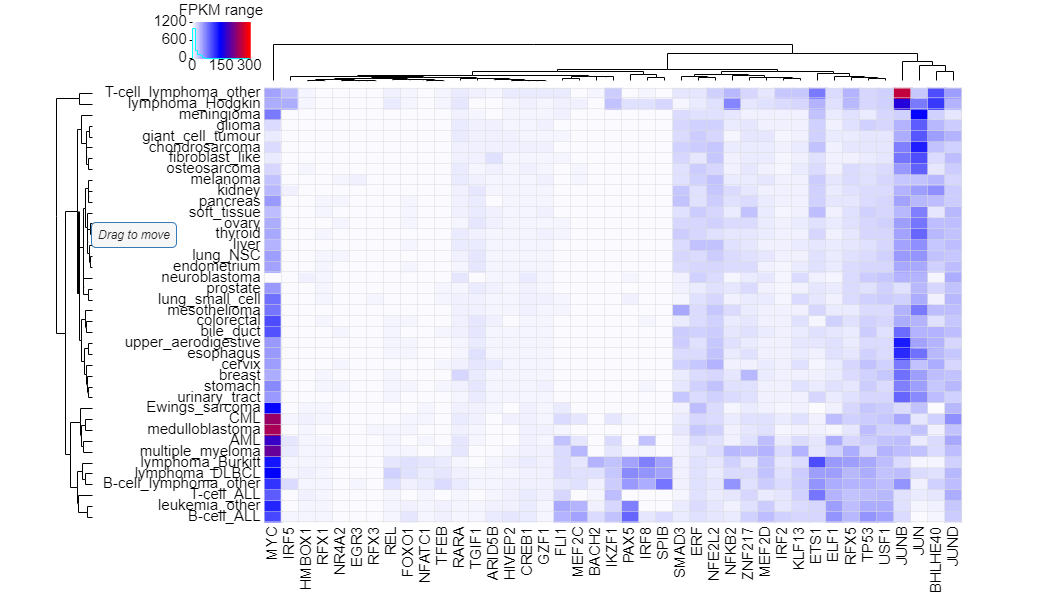
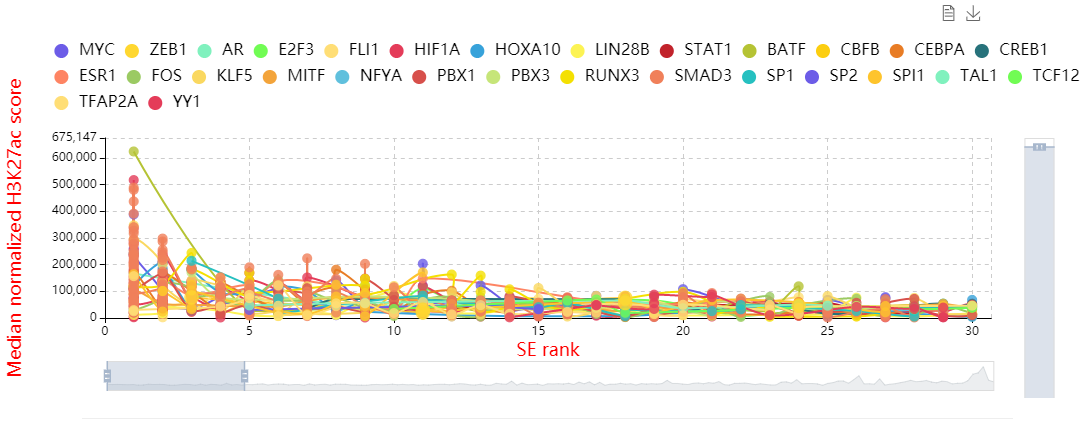
Here, we developed a comprehensive human core transcriptional regulatory circuit database (CRCdb), which aims to identify CRCs for various cell/tissue types. The current version of CRCdb documented over 500 samples from NCBI, ENCODE and Roadmap. For all samples, CRCs were identified by using a unified system environment, taking advantages of ‘coltron’ and ‘CRC mapper’. The number of human samples have been increased nearly three-fold compared with the existing database and the scale of data is five times larger than the existing database. Especially, the number of CRCs in CRCdb is 8,611,549. CRCdb further provides detailed information of CRCs (the most representative CRC and all other CRCs), supporting the display of frequency of TFs in CRCs and in-degree/out-degree of TFs.
Highlights of the CRCdb
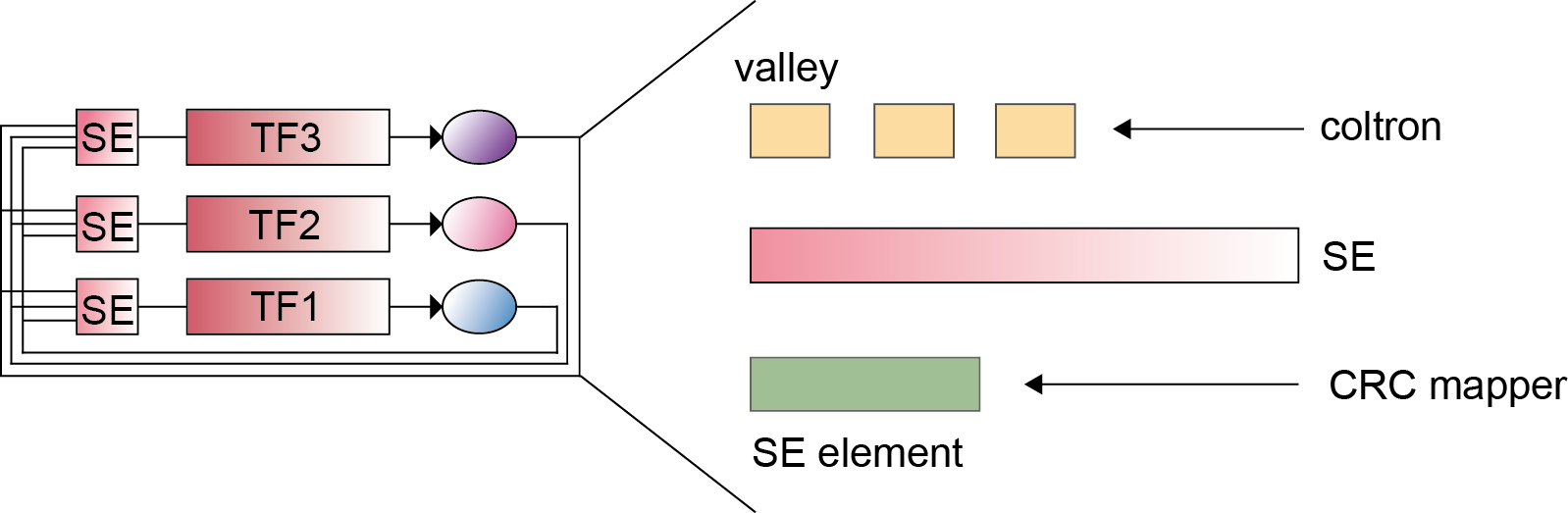
Two identification strategies
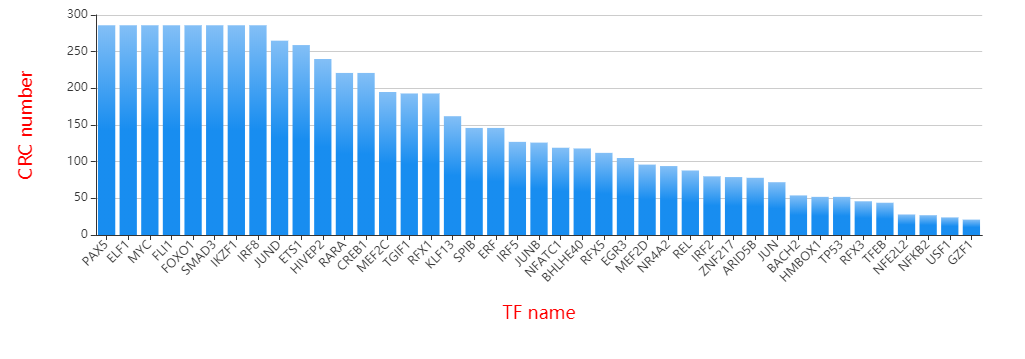
Frequency of TFs
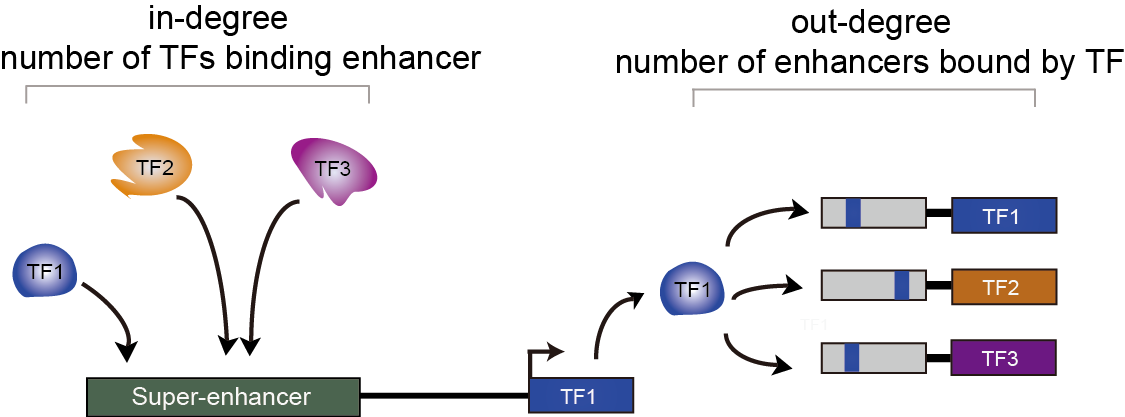
Degree of TFs
| Two identification strategy of CRCs. | Frequency of TFs that appear in CRCs. | Indegree and outdegree of each TF. |
| TF binding sites on SE regions. | The expression level of TF in tissues or cells. | TF families of TF (all CRCs). |
| Downstream regulation information of TF. | Genomic distribution of SEs associated with TF. | Mutation of TF. |
| Upstream regulation of TF. | Disease information of TF. | TF distribution in the most representative CRC. |
| TF distribution in all CRCs. |