1. What is Cancer CRC?
Core transcriptional regulatory circuit (CRC) is comprised of a group of interconnected auto-regulating transcription factors (TFs) forming loops, and it can be discovered by enhancer clusters called super-enhancers (SEs). Many studies have shown that CRCs play an important role in defining cellular identity, and core TFs in CRCs have been proved highly valuable for cell-type-specific transcriptional regulation in cancers.
Recently, a large number of cancer H3K27ac ChIP-seq data and ATAC-seq data have been accumulated rapidly, which promotes an urgent need for comprehensive and effective collecting and processing these data to obtain CRCs for various cancer types.
2. What information is available in the Cancer CRC?
Here, we developed a comprehensive cancer core transcriptional regulatory circuit platform (Cancer CRC), which aims to provide a large number of cancer CRCs, core TFs and the related significant information for core TFs in various cancers. The current version of Cancer CRC documented 94108 CRCs through using a ‘Coltron’ tool to integrate paired cancer ATAC-seq data and H3K27ac ChIP-seq data associated with specific cell lines. Importantly, the combination of two kinds data can identify the more precise TF-bound hypersensitive elements within large enhancer domains and SE-defined TF networks in different cancers than other methods. Cancer CRC platform also provides detailed annotation information of each CRC and core TF. Moreover, Cancer CRC provides ‘SE active core TFs analysis’ and ‘TF enrichment analysis’ tools to identify potentially important TFs in cancers.
3. Platform Content and Construction
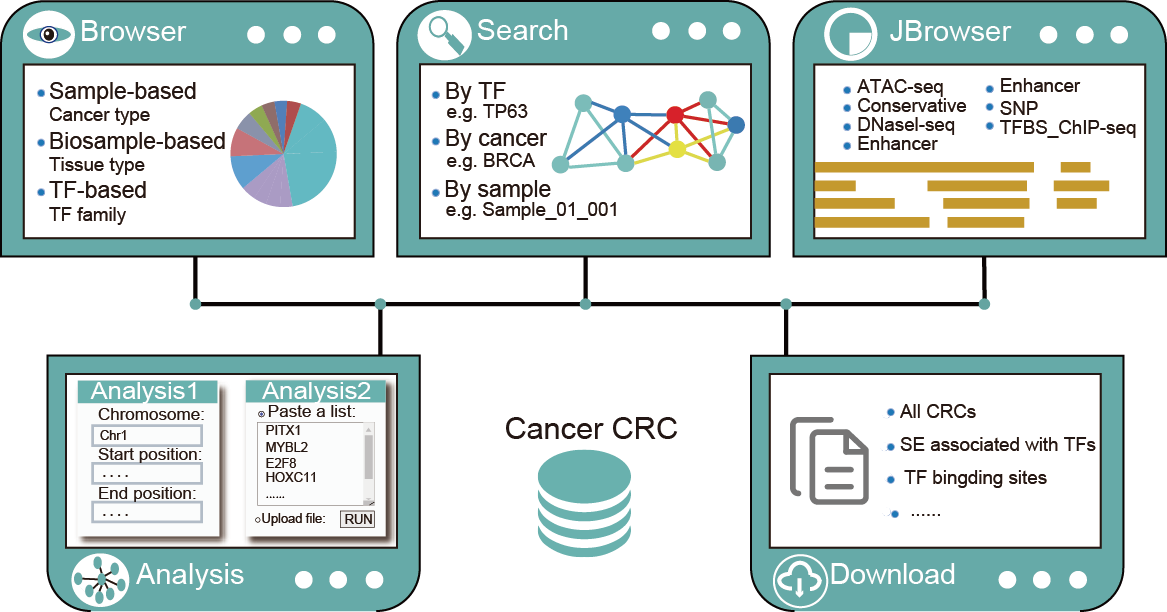
4.How to Use the Cancer CRC?
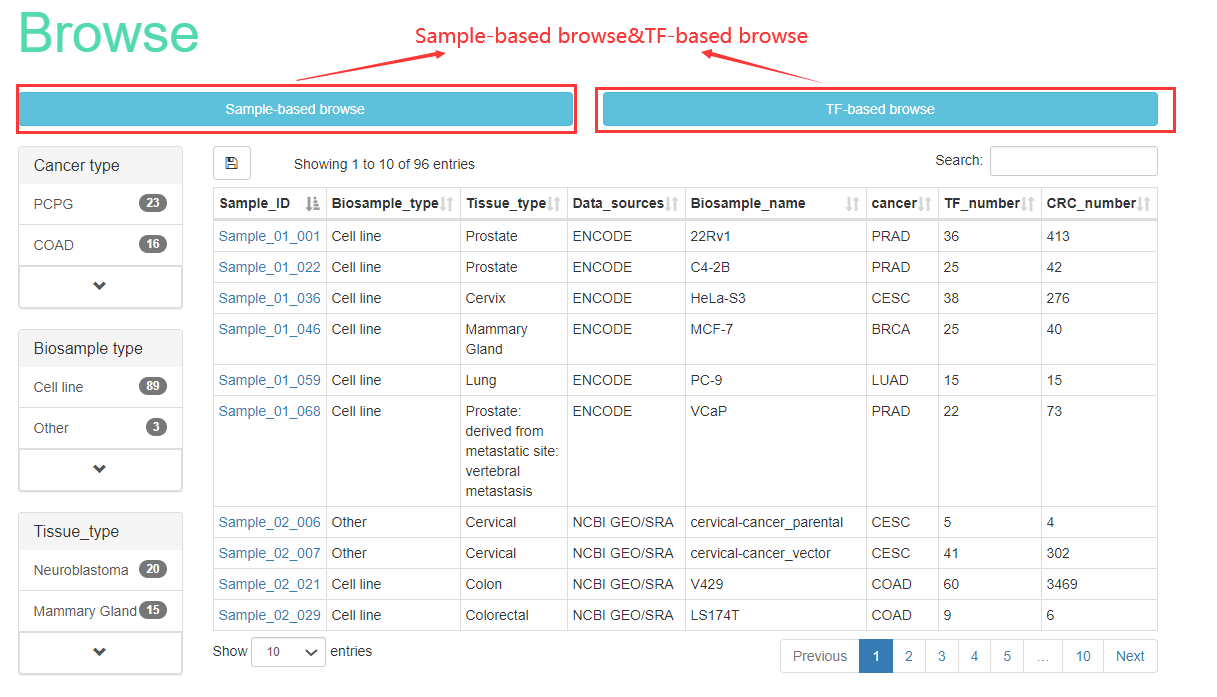
The ‘Browse’page is organized as an interactive and alphanumerically sortable table that allows users to quickly browse samples or TFs. Users can use the ‘Show entries’ drop-down menu to change the number of records per page. To view the CRCs for a given sample, users only need to click on the ‘Sample ID’ to view it. Besides, Cancer CRC also has TF-based browse. Users can browse related information about all TFs, which contains TF name, TF families, ensembl ID, entrez_ID and frequency of TFs.

Cancer CRC provides three query methods, including cancer-based, sample-based, and TF-based queries.
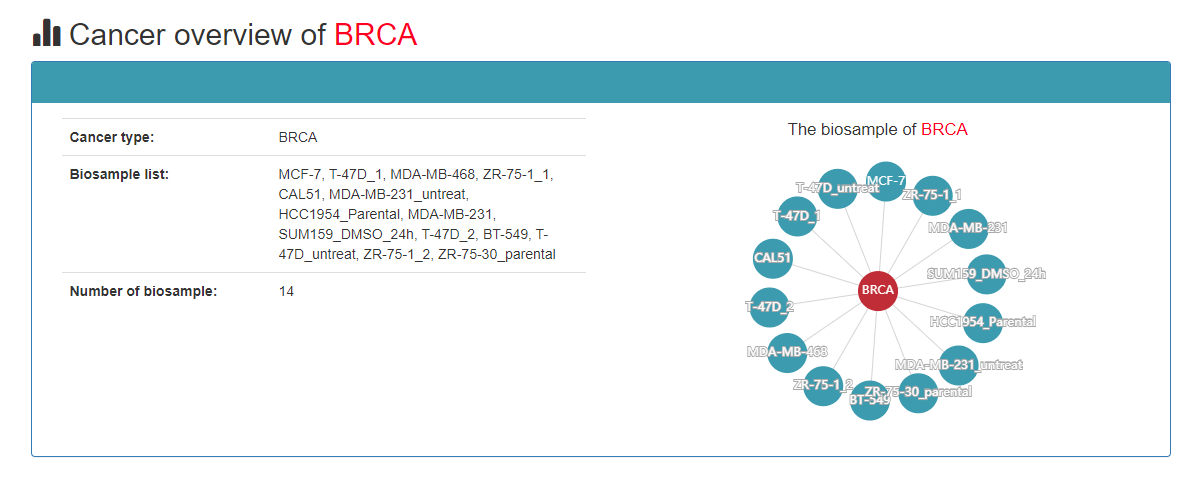
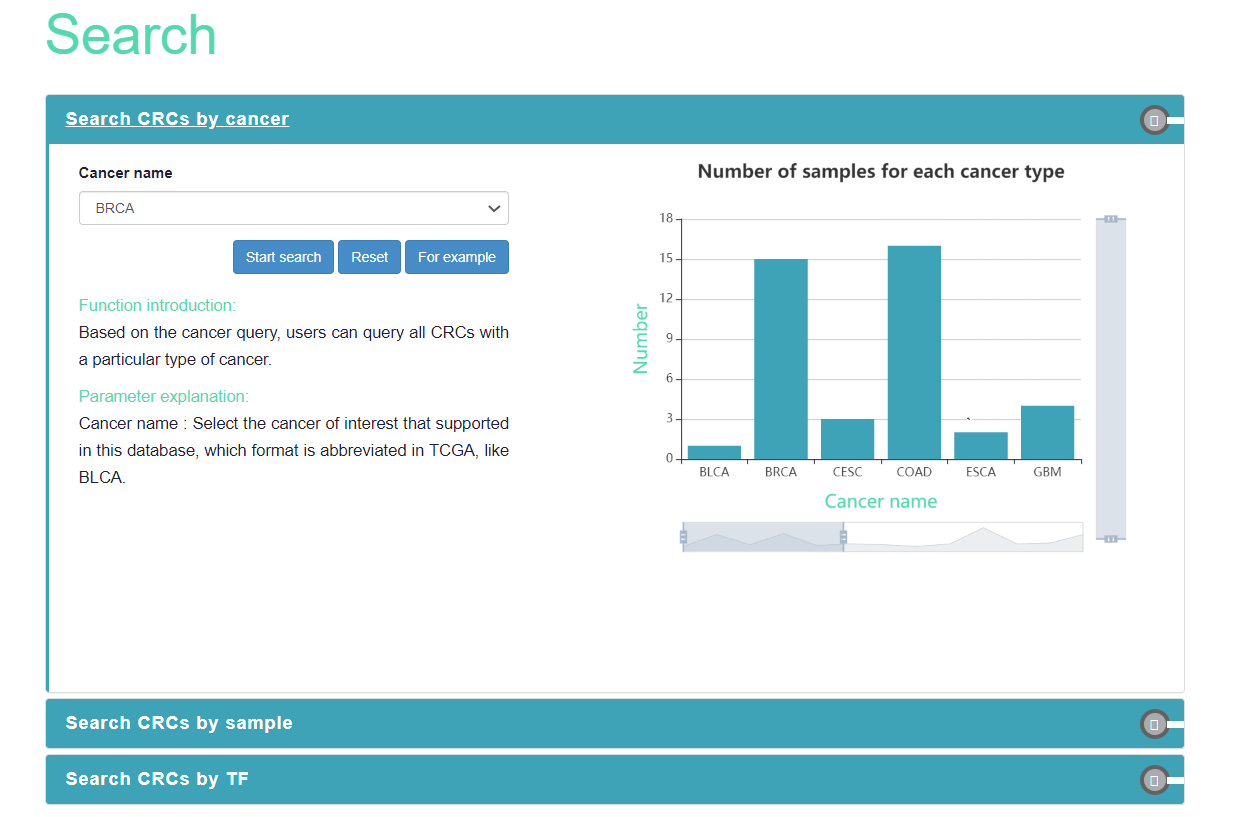 Based on the cancer type, users can query all CRCs with a particular type of cancer.
Based on the cancer type, users can query all CRCs with a particular type of cancer.
Users can see the biosamples of this cancer to further explore this cancer’s CRCs and these core TFs.
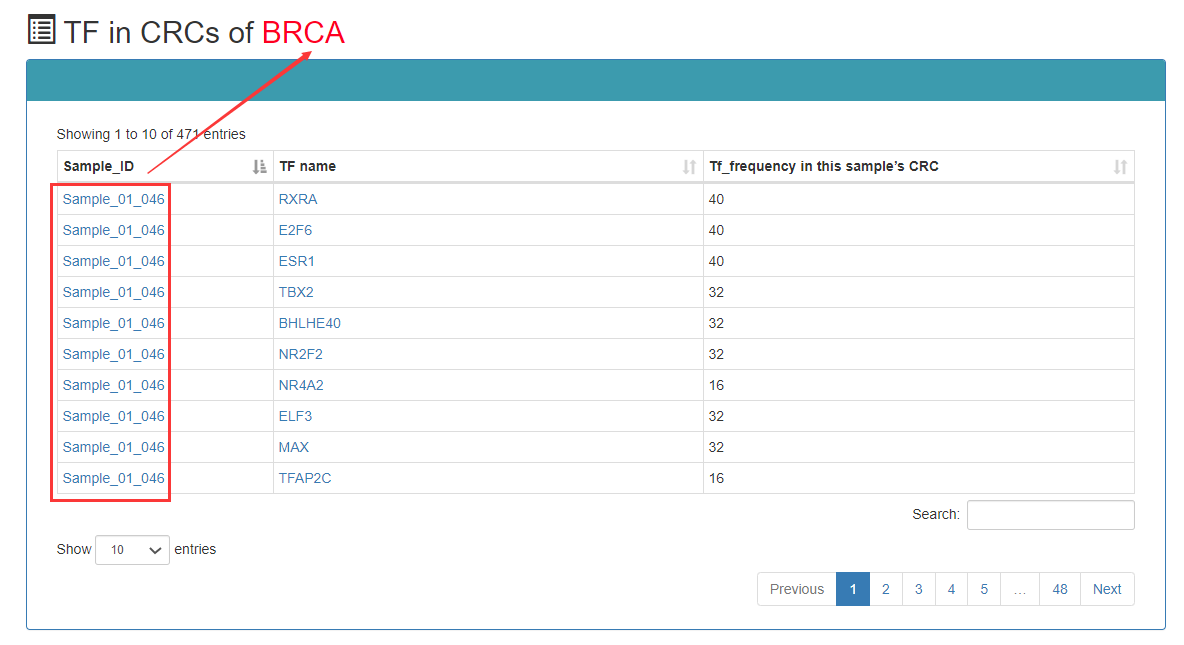 In the sample-based query, users can obtain the CRC result by determining a sample of the query.
In the sample-based query, users can obtain the CRC result by determining a sample of the query.
Users can see the most representative CRC and click on the TF to see the details. The most representative CRC is top ranked CRC.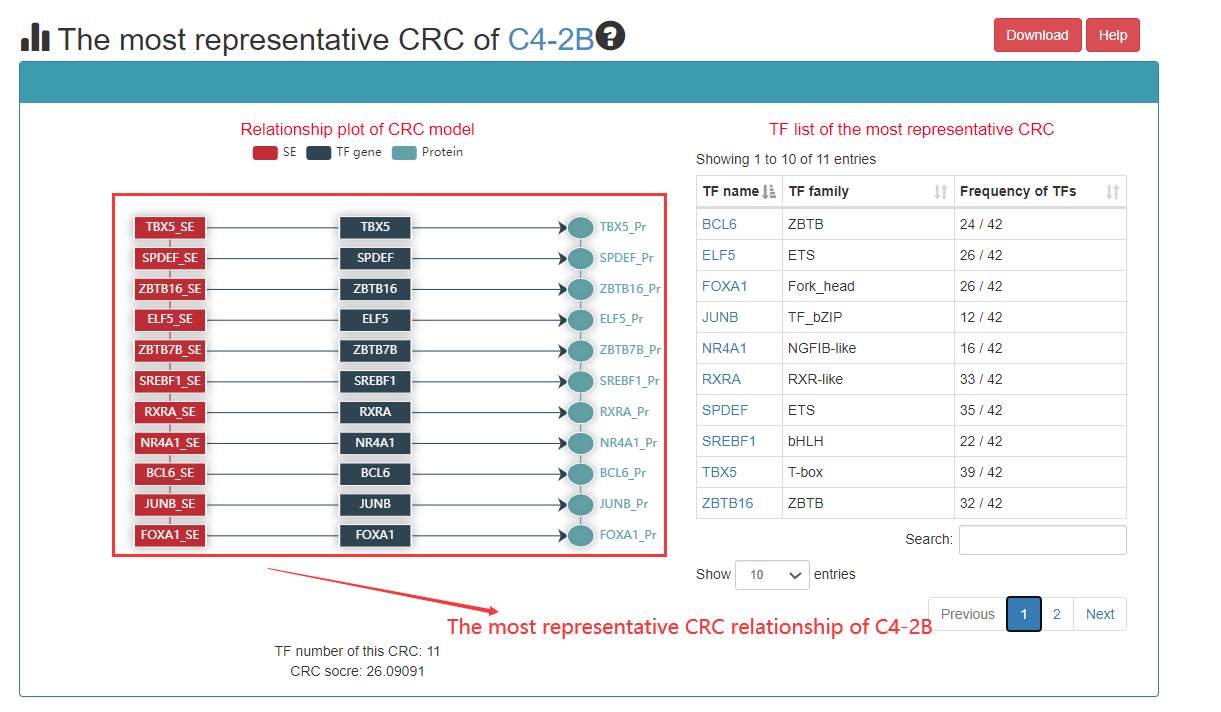
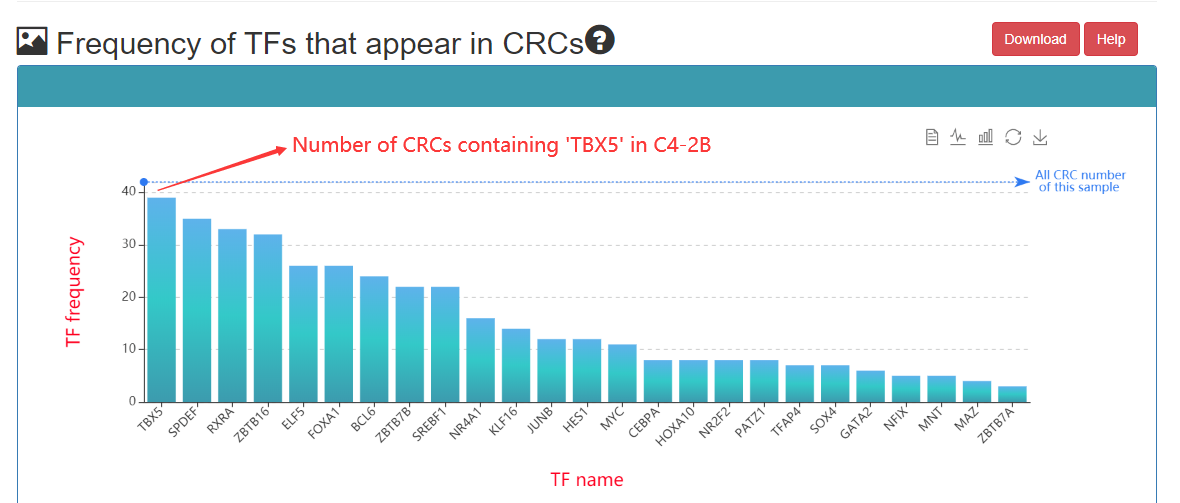
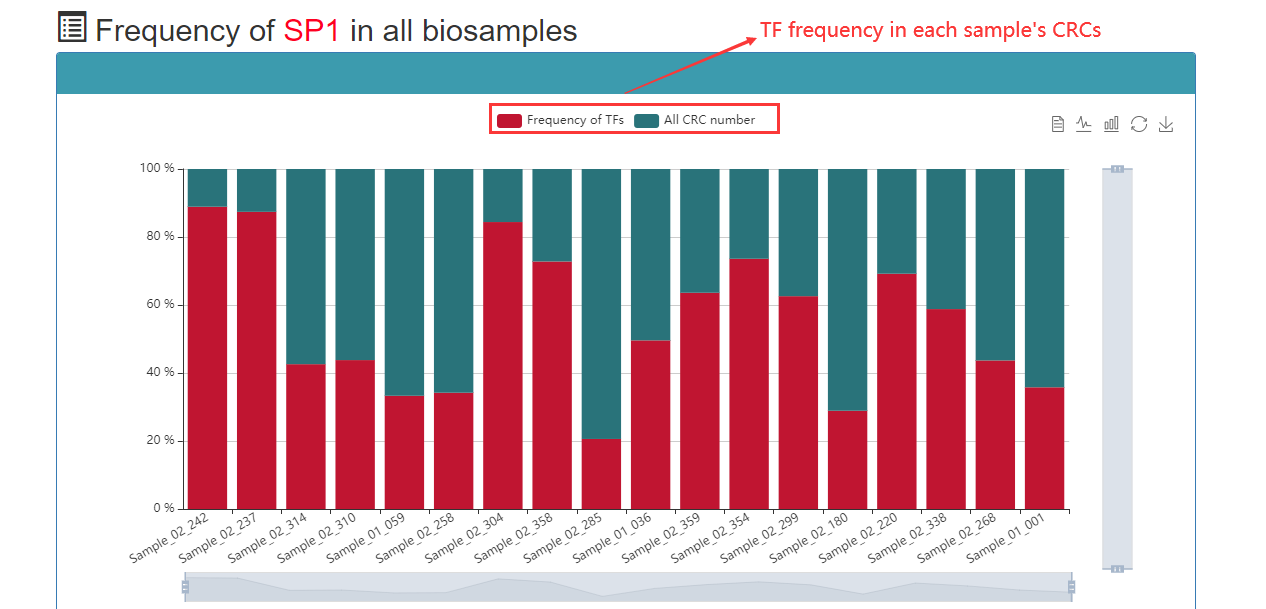
 Frequency of TFs that appear in CRCs: The greater the number of CRCs, the higher the frequency at which the TF is represented in all CRCs. Therefore, this TF may be more important under this sample.
Frequency of TFs that appear in CRCs: The greater the number of CRCs, the higher the frequency at which the TF is represented in all CRCs. Therefore, this TF may be more important under this sample.
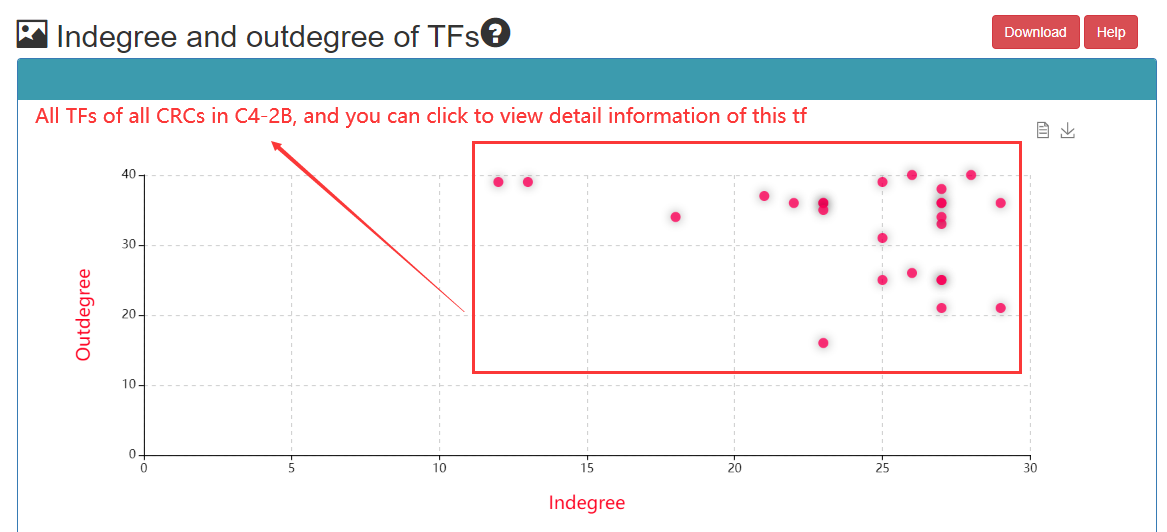
 Indegree: Number of TFs binding enhancer; Outdegree: Number of enhancers bound by TFs;
Indegree: Number of TFs binding enhancer; Outdegree: Number of enhancers bound by TFs;
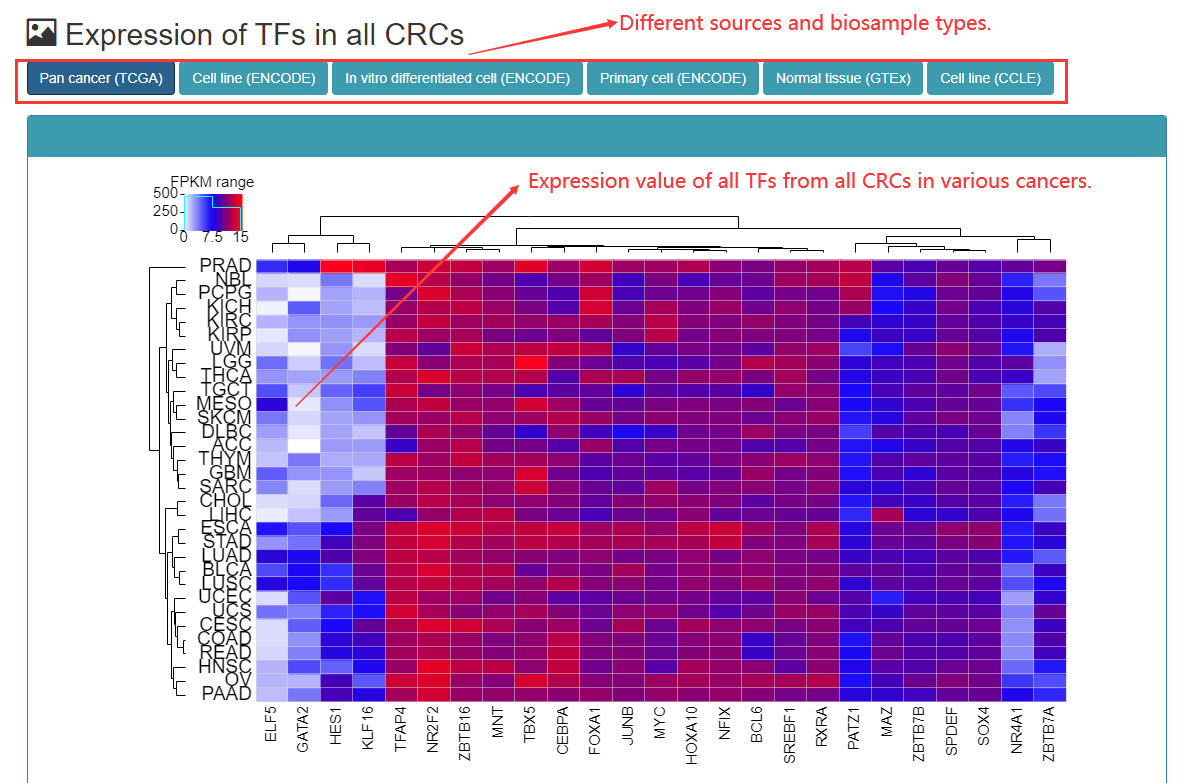
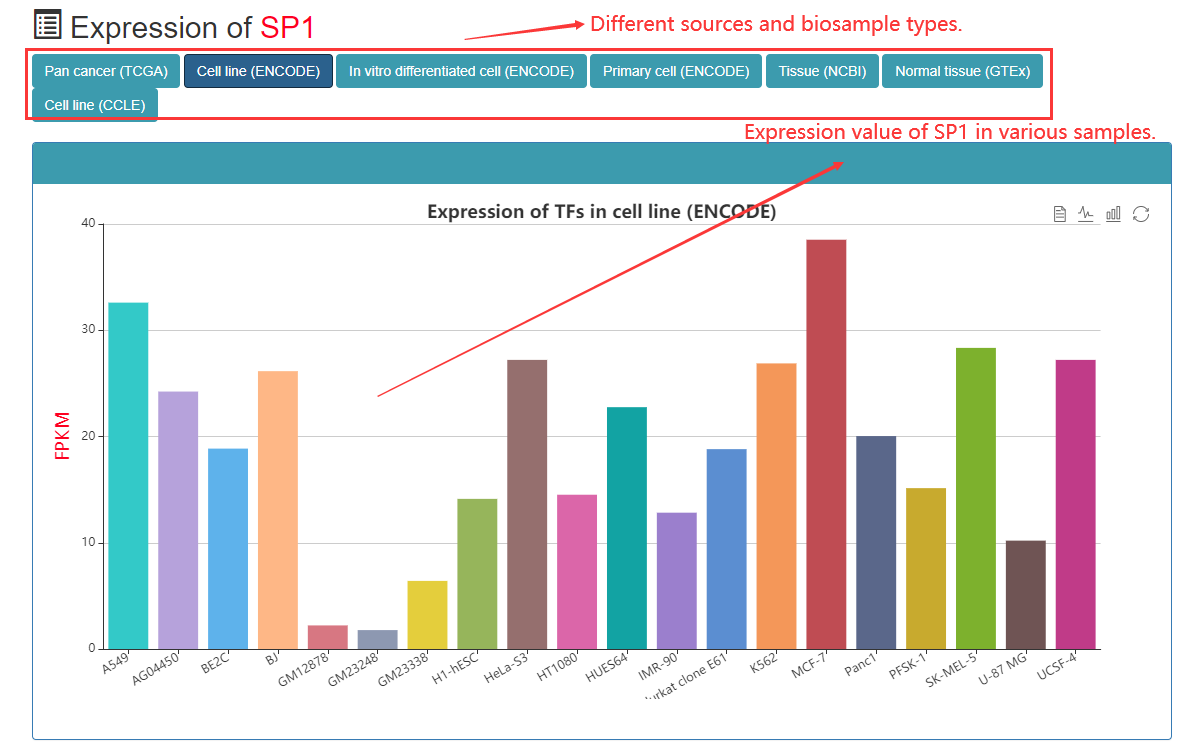
 Users can view the TF of the entire CRC of the sample, the amount of gene expression in tissues or cells. The expression level of TF is caculated using FPKM.
Users can view the TF of the entire CRC of the sample, the amount of gene expression in tissues or cells. The expression level of TF is caculated using FPKM.
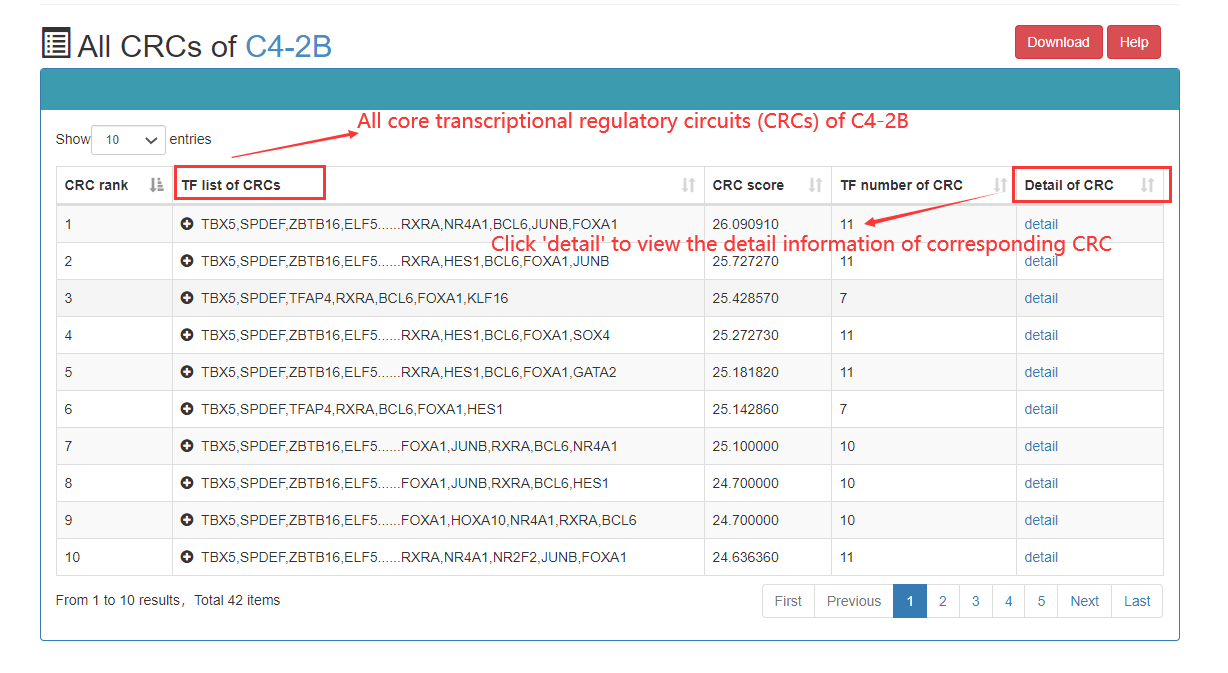
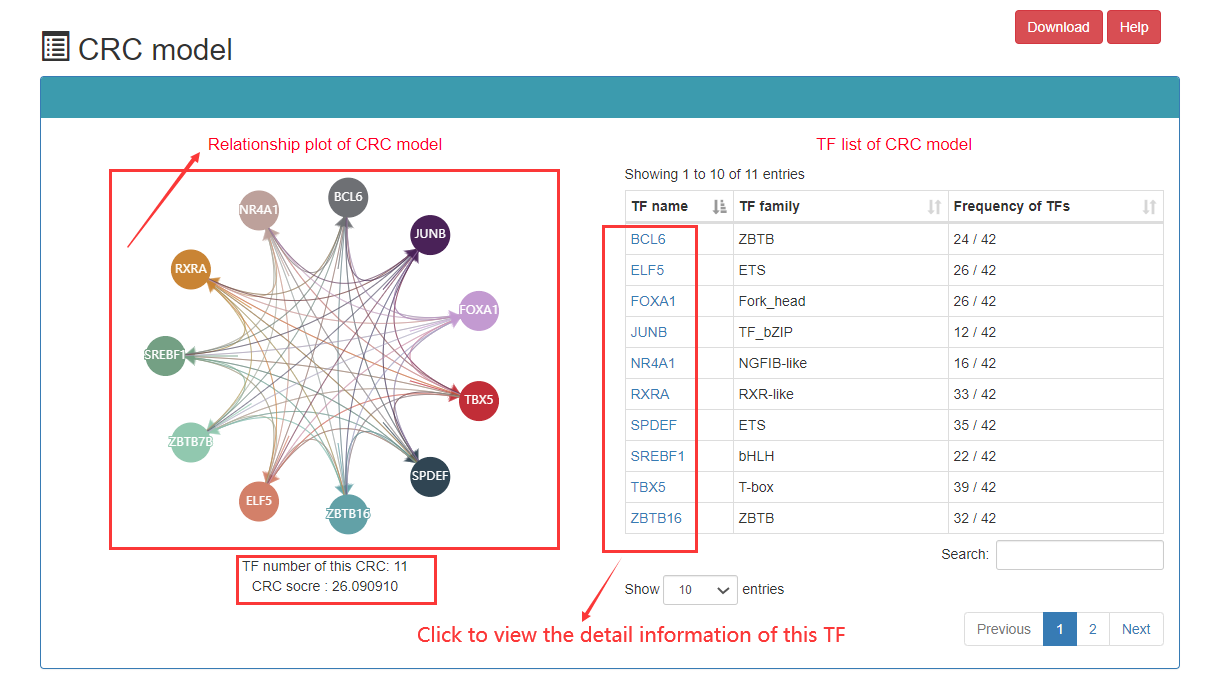
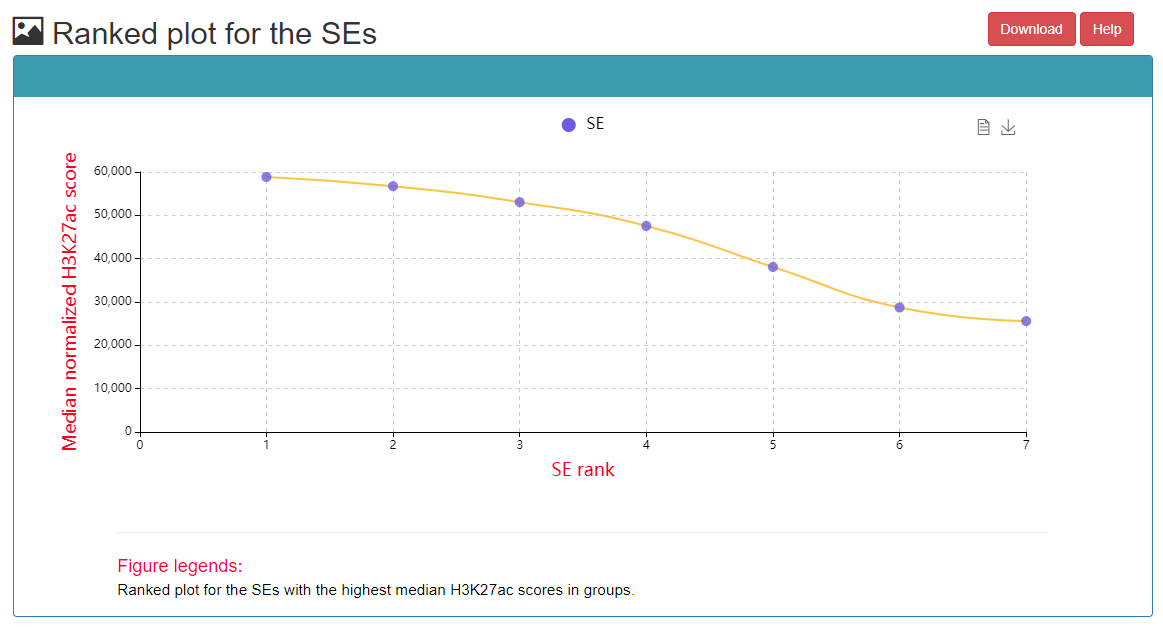
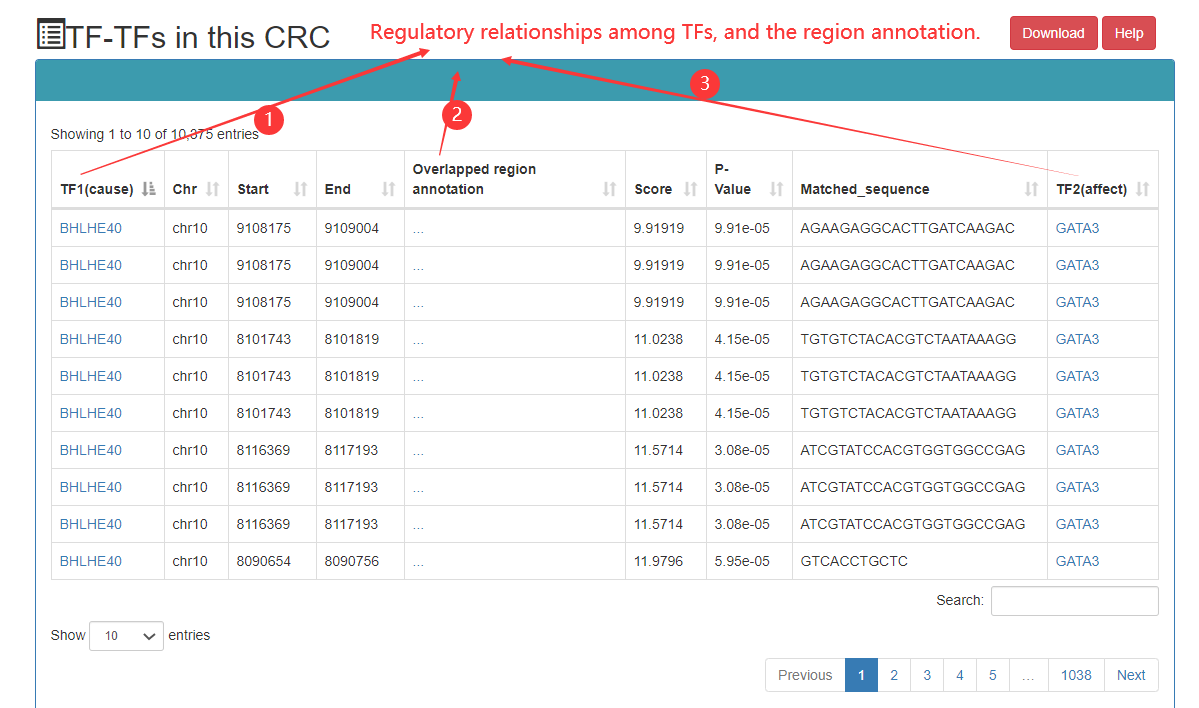
 In the CRC detailed page, the following information can got, including list of TF in this CRC, SE associated with core TFs, ranked plot for the SEs, regulatory relationships among TFs, etc.
In the CRC detailed page, the following information can got, including list of TF in this CRC, SE associated with core TFs, ranked plot for the SEs, regulatory relationships among TFs, etc.
 In the TF-based query, users can query a TF of interest, and then Cancer CRC will return all CRCs that match the TF–CRC relationship and distribute TF for all samples.
In the TF-based query, users can query a TF of interest, and then Cancer CRC will return all CRCs that match the TF–CRC relationship and distribute TF for all samples.
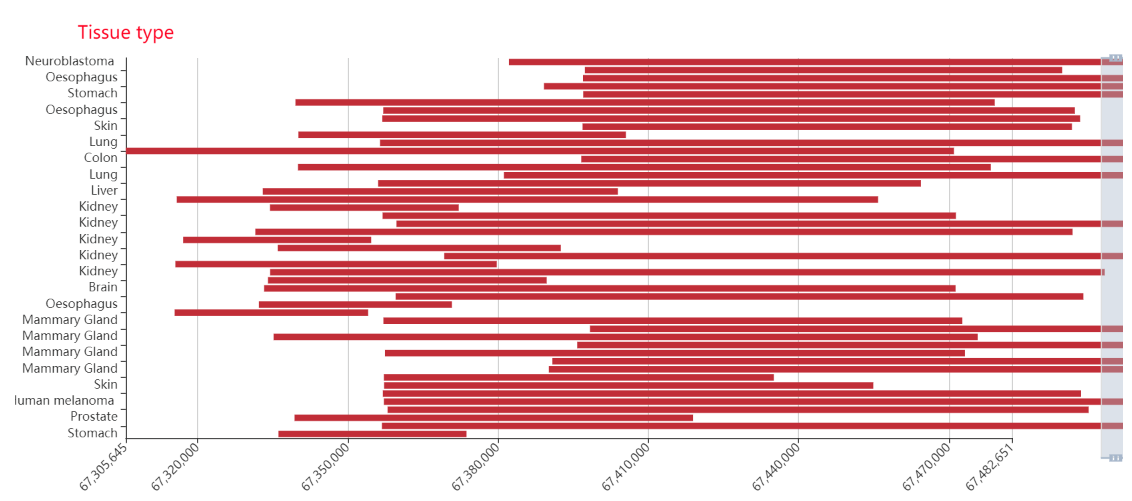
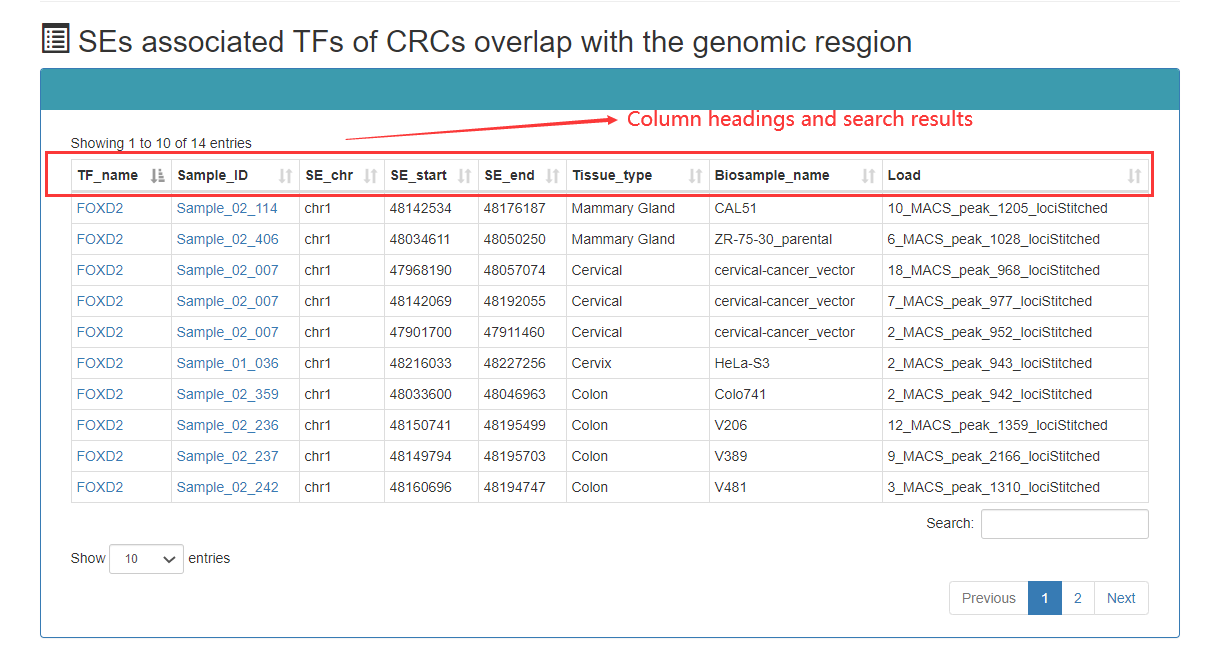
 Genomic distribution of SEs associated with TF: It can roughly understand the genomic regions of SEs associated with TF, and help to carry out the next analysis, such as knocking down the experiment.
Genomic distribution of SEs associated with TF: It can roughly understand the genomic regions of SEs associated with TF, and help to carry out the next analysis, such as knocking down the experiment.

Users can see the biosamples of this cancer to further explore this cancer’s CRCs and these core TFs.


Users can see the most representative CRC and click on the TF to see the details. The most representative CRC is top ranked CRC.












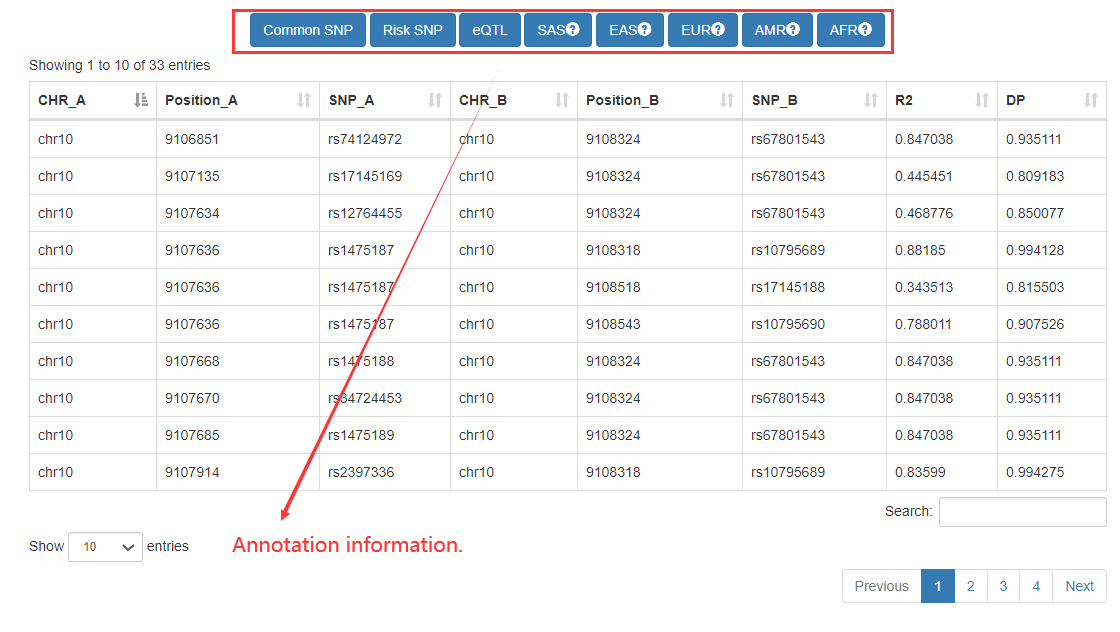
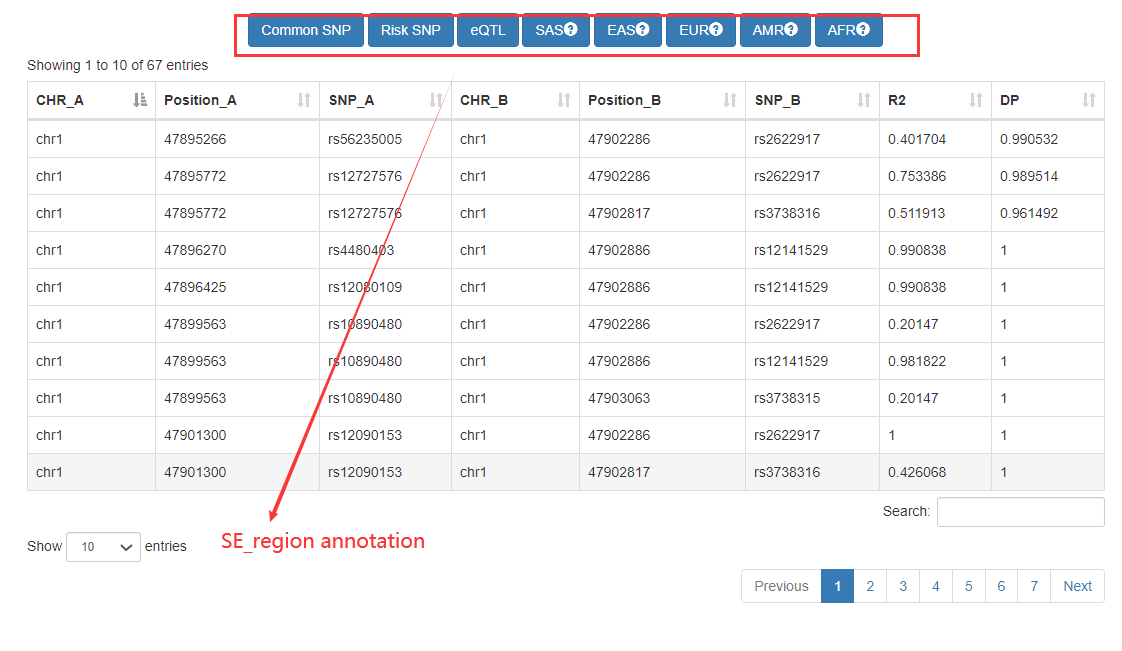
In the ‘Genomic-region-based’ analysis, Cancer CRC can identify SEs by inputting the genomic regions, and then find the core TFs associated with these SEs. For these identified SEs, we also provide detailed annotation information, including commom SNPs, risk SNPs, and eQTLs.
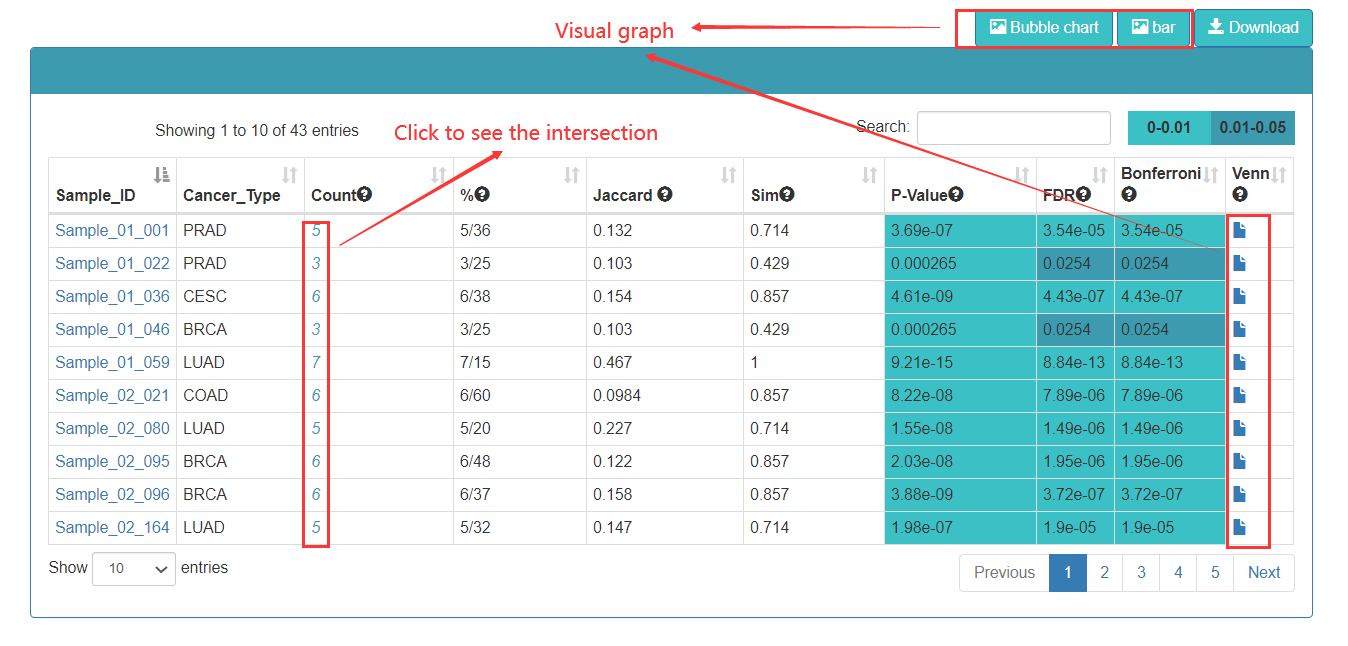
 In the TFs enrichment analysis, when users input an interesting TF list, Cancer CRC will annotate these TFs to reference TF sets, and calculate the statistical significance of enrichment analysis and similarity score. The result page displays the enrichment analysis results. Users can click on the top buttons to download the result table, and plot the enrichment analysis bubble chart and bar chart.
In the TFs enrichment analysis, when users input an interesting TF list, Cancer CRC will annotate these TFs to reference TF sets, and calculate the statistical significance of enrichment analysis and similarity score. The result page displays the enrichment analysis results. Users can click on the top buttons to download the result table, and plot the enrichment analysis bubble chart and bar chart.



The data of Cancer CRCs and the related files of all samples are available to download in the ‘Download’ page.

5.Frequently Asked Questions
Why might web pages load slowly?
Reply: Cancer CRC has advanced storage technology and sufficient bandwidth to meet the needs of most users for the speed of web page loading. However, it is not excluded that few users have poor user experience due to network reasons.What is the difference between the most representative CRC and all CRCs?
Reply: The most representative CRC is top ranked CRC; All CRCs is all CRCs identified by the identification strategy of CRCs
6.Development Environment
The current version of Cancer CRC was developed using MySQL 5.7.17 (http://www.mysql.com) and runs on a Linux-based Apache Web server (http://www.apache.org). We used PHP 5.6 (http://www.php.net) for server-side scripting. We designed and built the interactive interface using Bootstrap v3.3.7 (https://v3.bootcss.com) and JQuery v2.1.1 (http://jquery. com). We used ECharts (http://echarts.baidu.com) and D3 (https://d3js.org) as a graphical visualization framework, and JBrowse (http://jbrowse.org) as the genome browser framework. We recommend using a modern web browser that supports the HTML5 standard such as Firefox, Google Chrome, Safari and Opera or IE 9.0+ for the best display.
The Cancer CRC is freely available to the research community using the web link (http://bio.liclab.net/Cancer_crcdb/index.html). Users are not required to register or login to access features in the database.
The Cancer CRC is freely available to the research community using the web link (http://bio.liclab.net/Cancer_crcdb/index.html). Users are not required to register or login to access features in the database.