Welcome To Cancer CRC
A Comprehensive Cancer Core Transcriptional Regulatory Circuit Resource and Analysis Platform.
Introduction
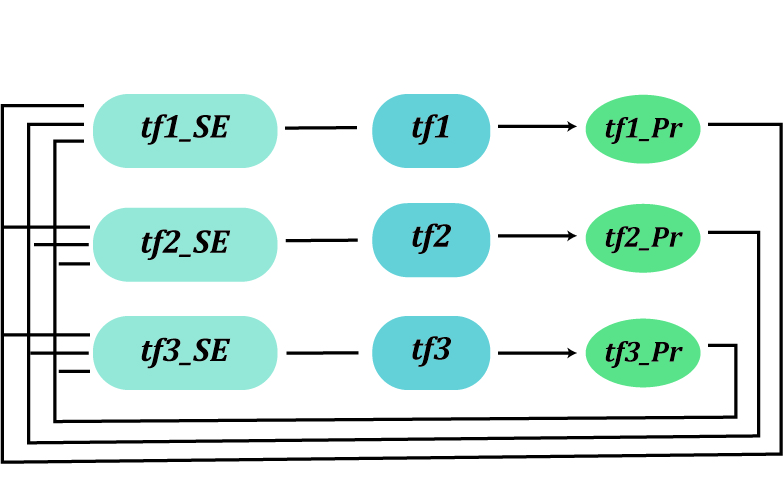
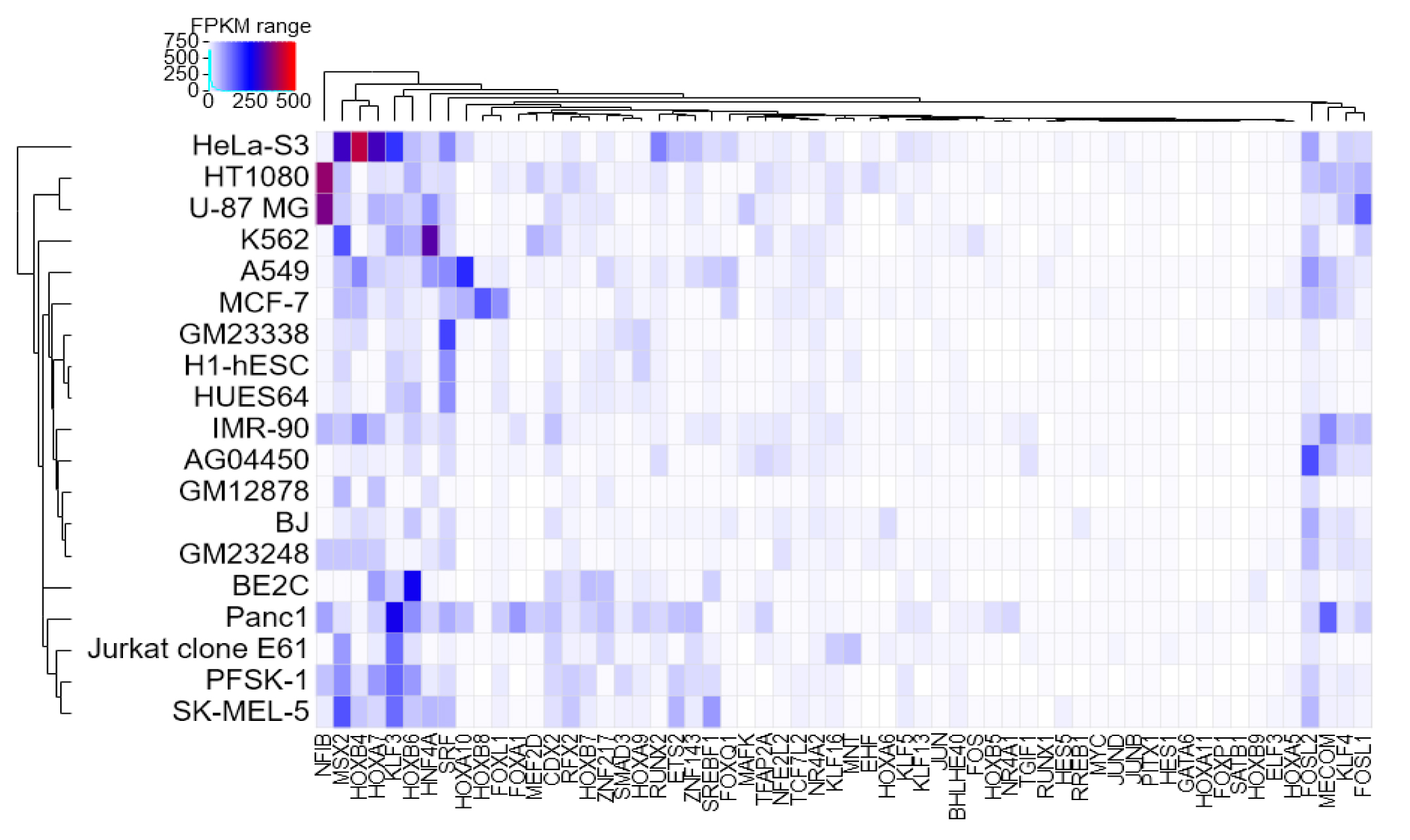
Core transcriptional regulatory circuit (CRC) is comprised of a group of interconnected auto-regulating transcription factors (TFs) forming loops, and it can be discovered by enhancer clusters called super-enhancers (SEs). Many studies have shown that CRCs play an important role in defining cellular identity, and core TFs in CRCs have been proved highly valuable for cell-type-specific transcriptional regulation in cancers.
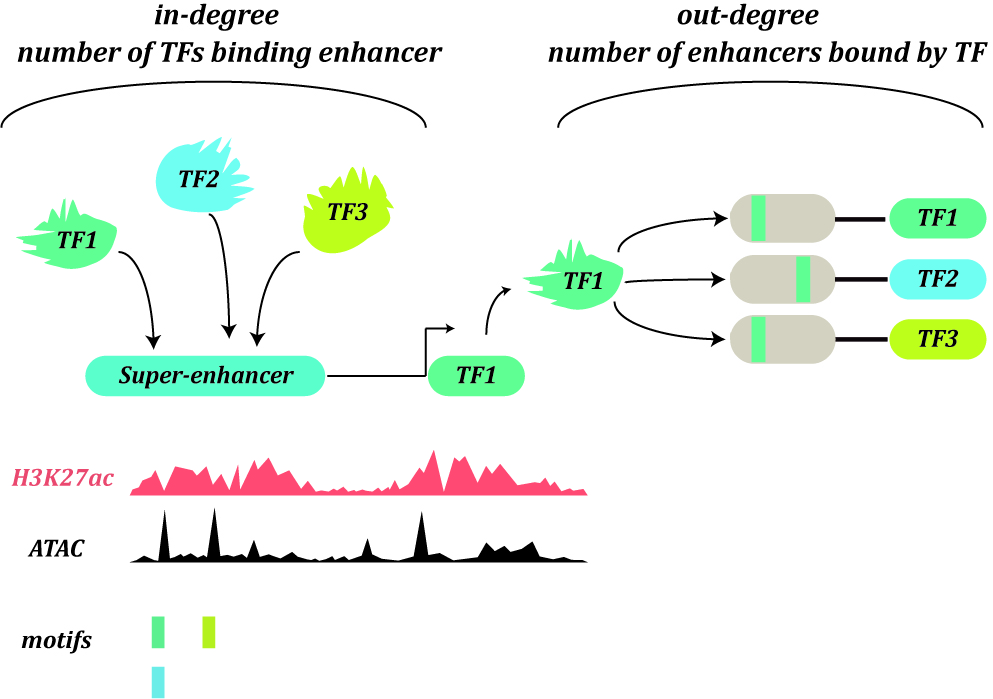
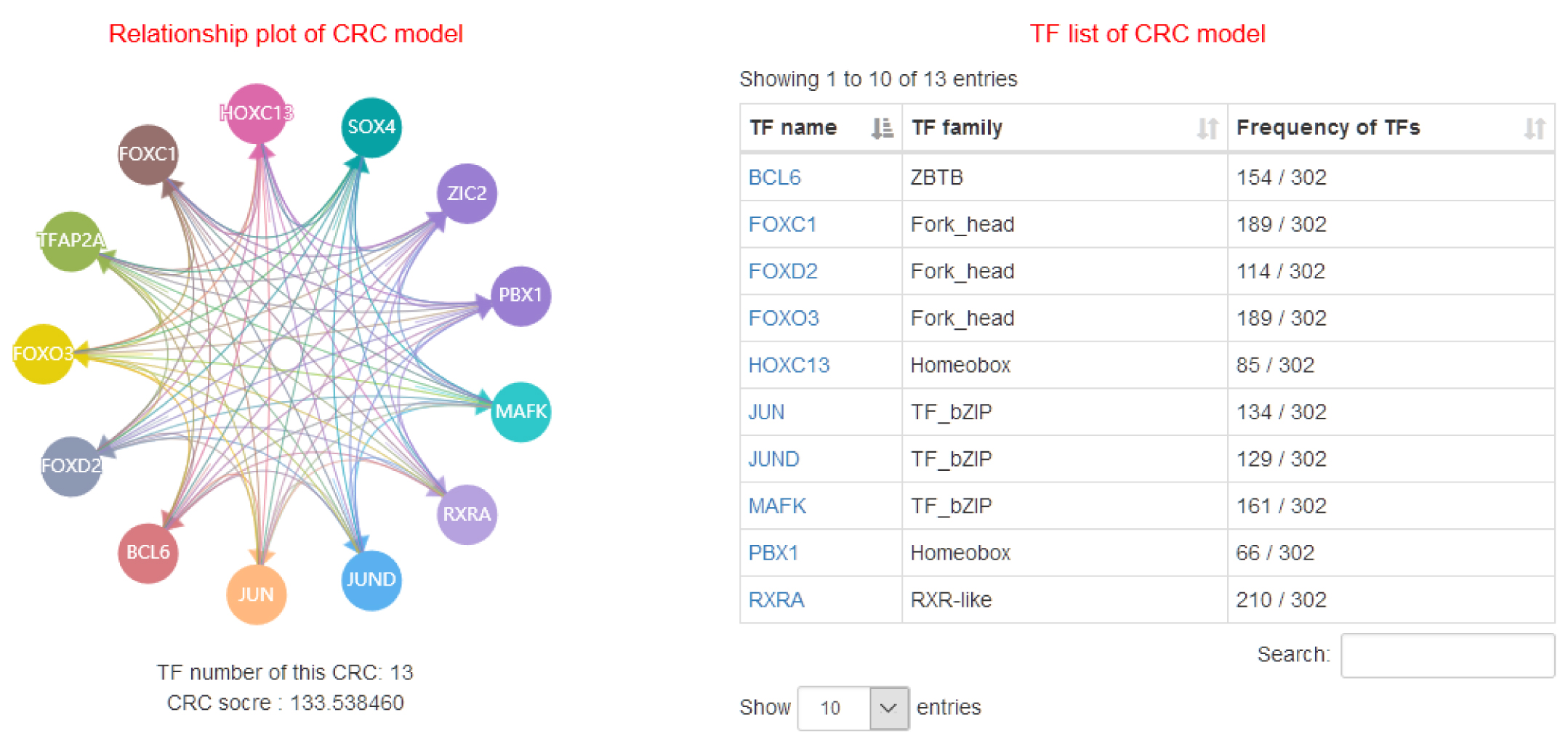
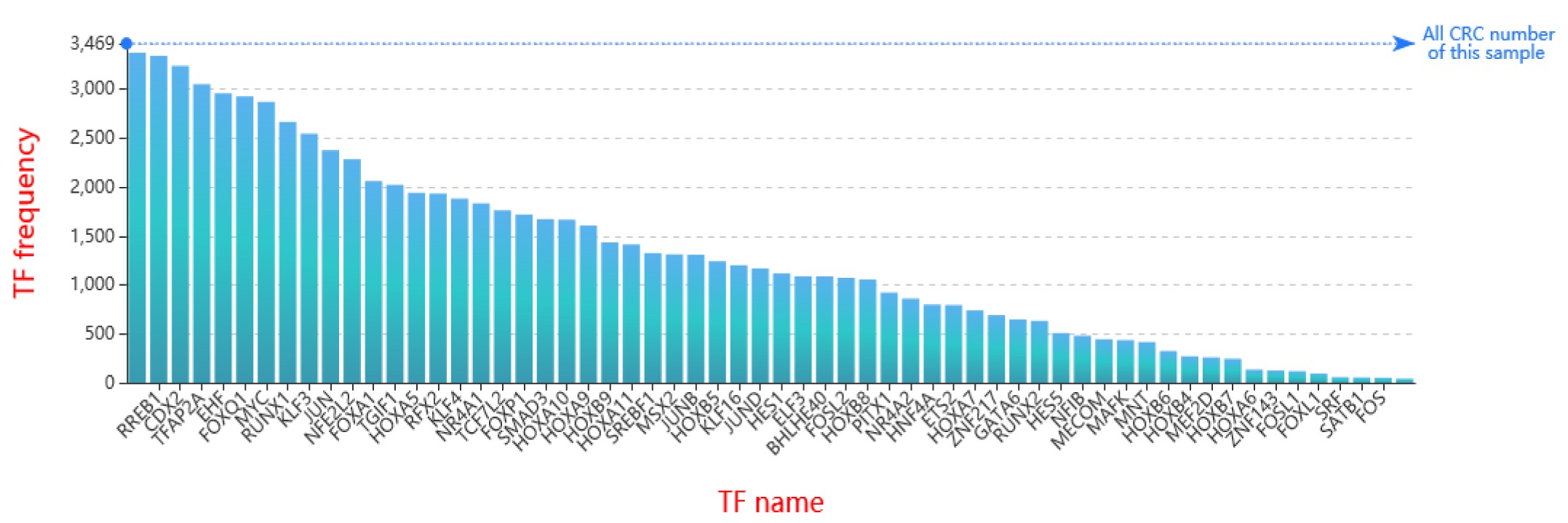
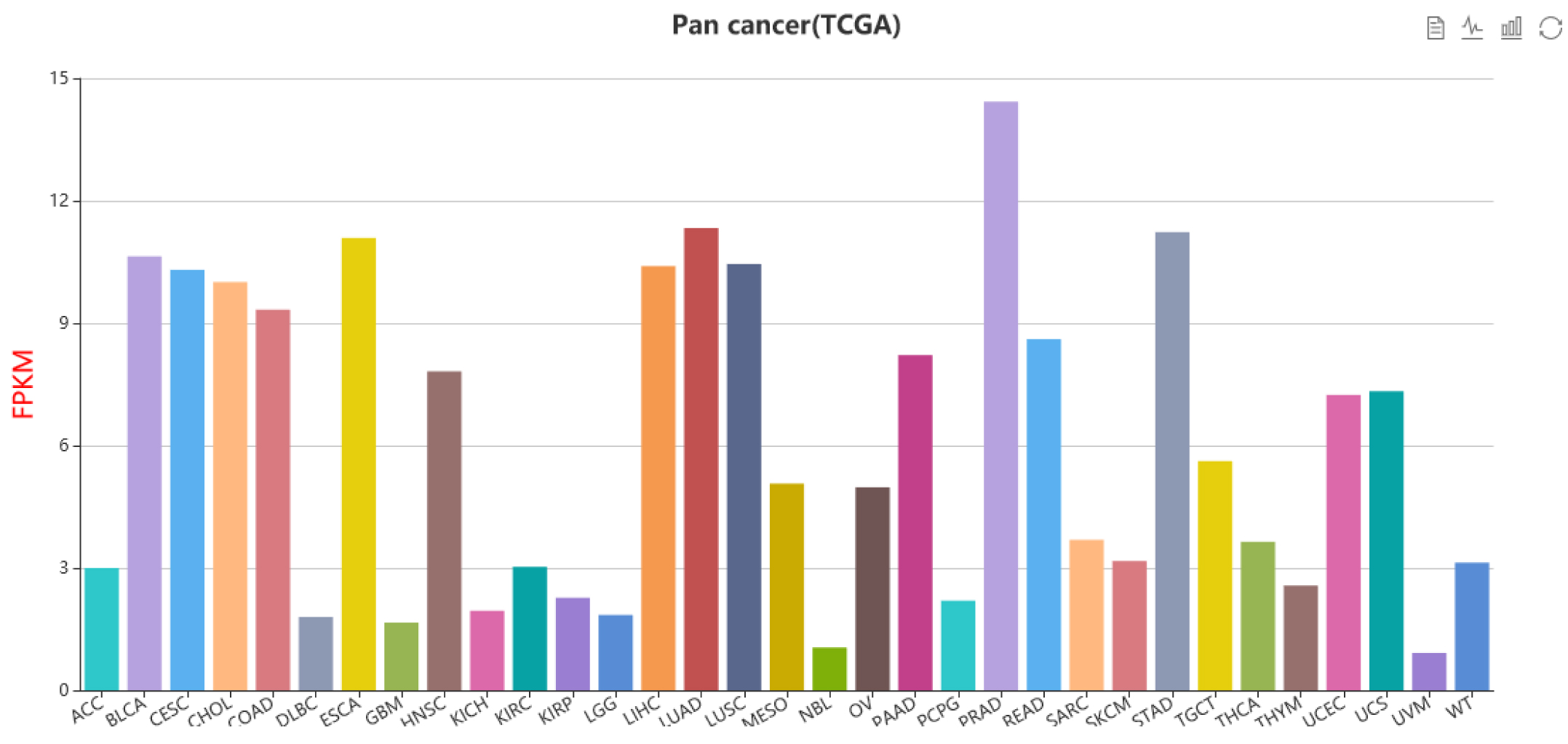
The current version of Cancer CRC documented 94,108 CRCs through using a ‘coltron’ tool to integrate many ATAC-seq and H3K27ac ChIP-seq data associated with specific cancer cell types. Importantly, Cancer CRC provides detailed annotation information of each CRC and core TFs, such as topological properties of TFs (in-degree and out-degree), expression value of TFs, frequency of core TFs in each sample of CRCs, SEs associated with TFs, cancer mutations of TFs, disease information, pathways associated with TFs, GO functions and survival information. Moreover, Cancer CRC supports enrichment analysis for TFs of interest. Cancer CRC provides a user-friendly interface to search, browse and visualize detailed information about these CRCs and core TFs.
Highlight of the Cancer CRC